+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30080 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the monomeric SPT-ORMDL3 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / lipid synthesis / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationsphinganine biosynthetic process / regulation of fat cell apoptotic process / negative regulation of ceramide biosynthetic process / sphingomyelin biosynthetic process / intracellular sphingolipid homeostasis / serine palmitoyltransferase complex / serine C-palmitoyltransferase activity / serine C-palmitoyltransferase / ceramide metabolic process / sphingosine biosynthetic process ...sphinganine biosynthetic process / regulation of fat cell apoptotic process / negative regulation of ceramide biosynthetic process / sphingomyelin biosynthetic process / intracellular sphingolipid homeostasis / serine palmitoyltransferase complex / serine C-palmitoyltransferase activity / serine C-palmitoyltransferase / ceramide metabolic process / sphingosine biosynthetic process / sphingolipid metabolic process / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / regulation of smooth muscle contraction / ceramide biosynthetic process / positive regulation of lipophagy / negative regulation of B cell apoptotic process / motor behavior / adipose tissue development / specific granule membrane / myelination / positive regulation of autophagy / secretory granule membrane / positive regulation of protein localization to nucleus / intracellular protein localization / pyridoxal phosphate binding / Neutrophil degranulation / endoplasmic reticulum membrane / endoplasmic reticulum / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Li SS / Xie T | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2021 Journal: Nat Struct Mol Biol / Year: 2021Title: Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex. Authors: Sisi Li / Tian Xie / Peng Liu / Lei Wang / Xin Gong /  Abstract: Human serine palmitoyltransferase (SPT) complex catalyzes the initial and rate-limiting step in the de novo biosynthesis of all sphingolipids. ORMDLs regulate SPT function, with human ORMDL3 being ...Human serine palmitoyltransferase (SPT) complex catalyzes the initial and rate-limiting step in the de novo biosynthesis of all sphingolipids. ORMDLs regulate SPT function, with human ORMDL3 being related to asthma. Here we report three high-resolution cryo-EM structures: the human SPT complex, composed of SPTLC1, SPTLC2 and SPTssa; the SPT-ORMDL3 complex; and the SPT-ORMDL3 complex bound to two substrates, PLP-L-serine (PLS) and a non-reactive palmitoyl-CoA analogue. SPTLC1 and SPTLC2 form a dimer of heterodimers as the catalytic core. SPTssa participates in acyl-CoA coordination, thereby stimulating the SPT activity and regulating the substrate selectivity. ORMDL3 is located in the center of the complex, serving to stabilize the SPT assembly. Our structural and biochemical analyses provide a molecular basis for the assembly and substrate selectivity of the SPT and SPT-ORMDL3 complexes, and lay a foundation for mechanistic understanding of sphingolipid homeostasis and for related therapeutic drug development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30080.map.gz emd_30080.map.gz | 116.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30080-v30.xml emd-30080-v30.xml emd-30080.xml emd-30080.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30080.png emd_30080.png | 43 KB | ||
| Filedesc metadata |  emd-30080.cif.gz emd-30080.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30080 http://ftp.pdbj.org/pub/emdb/structures/EMD-30080 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30080 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30080 | HTTPS FTP |
-Related structure data
| Related structure data |  6m4oMC  6m4nC  7cqiC  7cqkC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30080.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30080.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.114 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
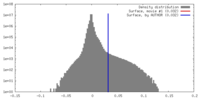
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : membrane protein 2
| Entire | Name: membrane protein 2 |
|---|---|
| Components |
|
-Supramolecule #1: membrane protein 2
| Supramolecule | Name: membrane protein 2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Serine palmitoyltransferase 1
| Macromolecule | Name: Serine palmitoyltransferase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: serine C-palmitoyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.80677 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATATEQWVL VEMVQALYEA PAYHLILEGI LILWIIRLLF SKTYKLQERS DLTVKEKEEL IEEWQPEPLV PPVPKDHPAL NYNIVSGPP SHKTVVNGKE CINFASFNFL GLLDNPRVKA AALASLKKYG VGTCGPRGFY GTFDVHLDLE DRLAKFMKTE E AIIYSYGF ...String: MATATEQWVL VEMVQALYEA PAYHLILEGI LILWIIRLLF SKTYKLQERS DLTVKEKEEL IEEWQPEPLV PPVPKDHPAL NYNIVSGPP SHKTVVNGKE CINFASFNFL GLLDNPRVKA AALASLKKYG VGTCGPRGFY GTFDVHLDLE DRLAKFMKTE E AIIYSYGF ATIASAIPAY SKRGDIVFVD RAACFAIQKG LQASRSDIKL FKHNDMADLE RLLKEQEIED QKNPRKARVT RR FIVVEGL YMNTGTICPL PELVKLKYKY KARIFLEESL SFGVLGEHGR GVTEHYGINI DDIDLISANM ENALASIGGF CCG RSFVID HQRLSGQGYC FSASLPPLLA AAAIEALNIM EENPGIFAVL KEKCGQIHKA LQGISGLKVV GESLSPAFHL QLEE STGSR EQDVRLLQEI VDQCMNRSIA LTQARYLEKE EKCLPPPSIR VVVTVEQTEE ELERAASTIK EVAQAVLL UniProtKB: Serine palmitoyltransferase 1 |
-Macromolecule #2: Serine palmitoyltransferase 2
| Macromolecule | Name: Serine palmitoyltransferase 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: serine C-palmitoyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 63.00416 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRPEPGGCCC RRTVRANGCV ANGEVRNGYV RSSAAAAAAA AAGQIHHVTQ NGGLYKRPFN EAFEETPMLV AVLTYVGYGV LTLFGYLRD FLRYWRIEKC HHATEREEQK DFVSLYQDFE NFYTRNLYMR IRDNWNRPIC SVPGARVDIM ERQSHDYNWS F KYTGNIIK ...String: MRPEPGGCCC RRTVRANGCV ANGEVRNGYV RSSAAAAAAA AAGQIHHVTQ NGGLYKRPFN EAFEETPMLV AVLTYVGYGV LTLFGYLRD FLRYWRIEKC HHATEREEQK DFVSLYQDFE NFYTRNLYMR IRDNWNRPIC SVPGARVDIM ERQSHDYNWS F KYTGNIIK GVINMGSYNY LGFARNTGSC QEAAAKVLEE YGAGVCSTRQ EIGNLDKHEE LEELVARFLG VEAAMAYGMG FA TNSMNIP ALVGKGCLIL SDELNHASLV LGARLSGATI RIFKHNNMQS LEKLLKDAIV YGQPRTRRPW KKILILVEGI YSM EGSIVR LPEVIALKKK YKAYLYLDEA HSIGALGPTG RGVVEYFGLD PEDVDVMMGT FTKSFGASGG YIGGKKELID YLRT HSHSA VYATSLSPPV VEQIITSMKC IMGQDGTSLG KECVQQLAEN TRYFRRRLKE MGFIIYGNED SPVVPLMLYM PAKIG AFGR EMLKRNIGVV VVGFPATPII ESRARFCLSA AHTKEILDTA LKEIDEVGDL LQLKYSRHRL VPLLDRPFDE TTYEET ED UniProtKB: Serine palmitoyltransferase 2 |
-Macromolecule #3: ORM1-like protein 3
| Macromolecule | Name: ORM1-like protein 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.512594 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNVGTAHSEV NPNTRVMNSR GIWLSYVLAI GLLHIVLLSI PFVSVPVVWT LTNLIHNMGM YIFLHTVKGT PFETPDQGKA RLLTHWEQM DYGVQFTASR KFLTITPIVL YFLTSFYTKY DQIHFVLNTV SLMSVLIPKL PQLHGVRIFG INKY UniProtKB: ORM1-like protein 3 |
-Macromolecule #4: Serine palmitoyltransferase small subunit A
| Macromolecule | Name: Serine palmitoyltransferase small subunit A / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.742409 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADYKDDDDK SGPDEVDASG RMAGMALARA WKQMSWFYYQ YLLVTALYML EPWERTVFNS MLVSIVGMAL YTGYVFMPQH IMAILHYFE IVQ UniProtKB: Serine palmitoyltransferase small subunit A |
-Macromolecule #5: PYRIDOXAL-5'-PHOSPHATE
| Macromolecule | Name: PYRIDOXAL-5'-PHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: PLP |
|---|---|
| Molecular weight | Theoretical: 247.142 Da |
| Chemical component information |  ChemComp-PLP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 59924 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)