[English] 日本語
 Yorodumi
Yorodumi- EMDB-30029: Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30029 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMiscellaneous transport and binding events / volume-sensitive anion channel activity / aspartate transmembrane transport / taurine transmembrane transport / monoatomic anion transmembrane transport / protein hexamerization / cellular response to osmotic stress / intracellular glucose homeostasis / monoatomic ion channel complex / intracellular signal transduction ...Miscellaneous transport and binding events / volume-sensitive anion channel activity / aspartate transmembrane transport / taurine transmembrane transport / monoatomic anion transmembrane transport / protein hexamerization / cellular response to osmotic stress / intracellular glucose homeostasis / monoatomic ion channel complex / intracellular signal transduction / endoplasmic reticulum membrane / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.36 Å | |||||||||
 Authors Authors | Nakamura R / Kasuya G | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2020 Journal: Commun Biol / Year: 2020Title: Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation. Authors: Ryoki Nakamura / Tomohiro Numata / Go Kasuya / Takeshi Yokoyama / Tomohiro Nishizawa / Tsukasa Kusakizako / Takafumi Kato / Tatsuya Hagino / Naoshi Dohmae / Masato Inoue / Kengo Watanabe / ...Authors: Ryoki Nakamura / Tomohiro Numata / Go Kasuya / Takeshi Yokoyama / Tomohiro Nishizawa / Tsukasa Kusakizako / Takafumi Kato / Tatsuya Hagino / Naoshi Dohmae / Masato Inoue / Kengo Watanabe / Hidenori Ichijo / Masahide Kikkawa / Mikako Shirouzu / Thomas J Jentsch / Ryuichiro Ishitani / Yasunobu Okada / Osamu Nureki /   Abstract: Members of the leucine-rich repeat-containing 8 (LRRC8) protein family, composed of the five LRRC8A-E isoforms, are pore-forming components of the volume-regulated anion channel (VRAC). LRRC8A and at ...Members of the leucine-rich repeat-containing 8 (LRRC8) protein family, composed of the five LRRC8A-E isoforms, are pore-forming components of the volume-regulated anion channel (VRAC). LRRC8A and at least one of the other LRRC8 isoforms assemble into heteromers to generate VRAC transport activities. Despite the availability of the LRRC8A structures, the structural basis of how LRRC8 isoforms other than LRRC8A contribute to the functional diversity of VRAC has remained elusive. Here, we present the structure of the human LRRC8D isoform, which enables the permeation of organic substrates through VRAC. The LRRC8D homo-hexamer structure displays a two-fold symmetric arrangement, and together with a structure-based electrophysiological analysis, revealed two key features. The pore constriction on the extracellular side is wider than that in the LRRC8A structures, which may explain the increased permeability of organic substrates. Furthermore, an N-terminal helix protrudes into the pore from the intracellular side and may be critical for gating. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30029.map.gz emd_30029.map.gz | 6.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30029-v30.xml emd-30029-v30.xml emd-30029.xml emd-30029.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30029.png emd_30029.png | 146.9 KB | ||
| Masks |  emd_30029_msk_1.map emd_30029_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-30029.cif.gz emd-30029.cif.gz | 6.6 KB | ||
| Others |  emd_30029_half_map_1.map.gz emd_30029_half_map_1.map.gz emd_30029_half_map_2.map.gz emd_30029_half_map_2.map.gz | 49.5 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30029 http://ftp.pdbj.org/pub/emdb/structures/EMD-30029 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30029 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30029 | HTTPS FTP |
-Validation report
| Summary document |  emd_30029_validation.pdf.gz emd_30029_validation.pdf.gz | 646.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30029_full_validation.pdf.gz emd_30029_full_validation.pdf.gz | 646.1 KB | Display | |
| Data in XML |  emd_30029_validation.xml.gz emd_30029_validation.xml.gz | 11.6 KB | Display | |
| Data in CIF |  emd_30029_validation.cif.gz emd_30029_validation.cif.gz | 13.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30029 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30029 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30029 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30029 | HTTPS FTP |
-Related structure data
| Related structure data |  6m04MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10383 (Title: CryoEM structure of human LRRC8D / Data size: 1.7 TB EMPIAR-10383 (Title: CryoEM structure of human LRRC8D / Data size: 1.7 TBData #1: Unaligned movies of human LRRC8D isoform [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30029.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30029.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.49 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
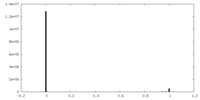
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_30029_msk_1.map emd_30029_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
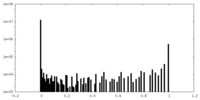
| Density Histograms |
-Half map: #1
| File | emd_30029_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
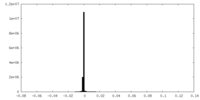
| Density Histograms |
-Half map: #2
| File | emd_30029_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
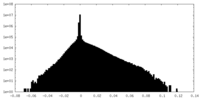
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric channel of LRC8D_HUMAN
| Entire | Name: Hexameric channel of LRC8D_HUMAN |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric channel of LRC8D_HUMAN
| Supramolecule | Name: Hexameric channel of LRC8D_HUMAN / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Volume-regulated anion channel subunit LRRC8D
| Macromolecule | Name: Volume-regulated anion channel subunit LRRC8D / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 99.31118 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFTLAEVASL NDIQPTYRIL KPWWDVFMDY LAVVMLMVAI FAGTMQLTKD QVVCLPVLPS PVNSKAHTPP GNAEVTTNIP KMEAATNQD QDGRTTNDIS FGTSAVTPDI PLRATYPRTD FALPNQEAKK EKKDPTGRKT NLDFQQYVFI NQMCYHLALP W YSKYFPYL ...String: MFTLAEVASL NDIQPTYRIL KPWWDVFMDY LAVVMLMVAI FAGTMQLTKD QVVCLPVLPS PVNSKAHTPP GNAEVTTNIP KMEAATNQD QDGRTTNDIS FGTSAVTPDI PLRATYPRTD FALPNQEAKK EKKDPTGRKT NLDFQQYVFI NQMCYHLALP W YSKYFPYL ALIHTIILMV SSNFWFKYPK TCSKVEHFVS ILGKCFESPW TTKALSETAC EDSEENKQRI TGAQTLPKHV ST SSDEGSP SASTPMINKT GFKFSAEKPV IEVPSMTILD KKDGEQAKAL FEKVRKFRAH VEDSDLIYKL YVVQTVIKTA KFI FILCYT ANFVNAISFE HVCKPKVEHL IGYEVFECTH NMAYMLKKLL ISYISIICVY GFICLYTLFW LFRIPLKEYS FEKV REESS FSDIPDVKND FAFLLHMVDQ YDQLYSKRFG VFLSEVSENK LREISLNHEW TFEKLRQHIS RNAQDKQELH LFMLS GVPD AVFDLTDLDV LKLELIPEAK IPAKISQMTN LQELHLCHCP AKVEQTAFSF LRDHLRCLHV KFTDVAEIPA WVYLLK NLR ELYLIGNLNS ENNKMIGLES LRELRHLKIL HVKSNLTKVP SNITDVAPHL TKLVIHNDGT KLLVLNSLKK MMNVAEL EL QNCELERIPH AIFSLSNLQE LDLKSNNIRT IEEIISFQHL KRLTCLKLWH NKIVTIPPSI THVKNLESLY FSNNKLES L PVAVFSLQKL RCLDVSYNNI SMIPIEIGLL QNLQHLHITG NKVDILPKQL FKCIKLRTLN LGQNCITSLP EKVGQLSQL TQLELKGNCF DRLPAQLGQC RMLKKSGLVV EDHLFDTLPL EVKEALNQDI NIPFANGIGT ENLYFQ UniProtKB: Volume-regulated anion channel subunit LRRC8D |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: The solution was freshly prepared to avoid digitonin precipitation. | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER/RHODIUM / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blotted for 4 seconds before plunging.. | |||||||||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 79.55 K / Max: 79.55 K |
| Details | Specimen holder is FEI Talos Arctica autogrid holder. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number real images: 3397 / Average exposure time: 15.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 23500 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6m04: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)