[English] 日本語
 Yorodumi
Yorodumi- EMDB-29926: Domoate-bound GluK2 kainate receptor in partially-open conformation 1 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Domoate-bound GluK2 kainate receptor in partially-open conformation 1 | |||||||||
 Map data Map data | Domoate-bound GluK2 kainate receptor in partially-open conformation 1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / glutamate kainate receptor 2 / homotetramer / MEMBRANE PROTEIN / partial agonist / gating mechanism | |||||||||
| Function / homology |  Function and homology information Function and homology informationmossy fiber rosette / detection of cold stimulus involved in thermoception / Activation of Na-permeable kainate receptors / Activation of Ca-permeable Kainate Receptor / kainate selective glutamate receptor complex / regulation of short-term neuronal synaptic plasticity / glutamate receptor activity / ubiquitin conjugating enzyme binding / negative regulation of synaptic transmission, glutamatergic / regulation of JNK cascade ...mossy fiber rosette / detection of cold stimulus involved in thermoception / Activation of Na-permeable kainate receptors / Activation of Ca-permeable Kainate Receptor / kainate selective glutamate receptor complex / regulation of short-term neuronal synaptic plasticity / glutamate receptor activity / ubiquitin conjugating enzyme binding / negative regulation of synaptic transmission, glutamatergic / regulation of JNK cascade / inhibitory postsynaptic potential / receptor clustering / kainate selective glutamate receptor activity / extracellularly glutamate-gated ion channel activity / modulation of excitatory postsynaptic potential / ionotropic glutamate receptor complex / positive regulation of synaptic transmission / behavioral fear response / neuronal action potential / glutamate-gated receptor activity / glutamate-gated calcium ion channel activity / dendrite cytoplasm / presynaptic modulation of chemical synaptic transmission / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / hippocampal mossy fiber to CA3 synapse / SNARE binding / PDZ domain binding / excitatory postsynaptic potential / regulation of membrane potential / synaptic transmission, glutamatergic / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / regulation of long-term neuronal synaptic plasticity / postsynaptic density membrane / modulation of chemical synaptic transmission / intracellular calcium ion homeostasis / terminal bouton / positive regulation of neuron apoptotic process / presynaptic membrane / neuron apoptotic process / scaffold protein binding / chemical synaptic transmission / negative regulation of neuron apoptotic process / perikaryon / postsynaptic membrane / postsynaptic density / axon / neuronal cell body / ubiquitin protein ligase binding / dendrite / synapse / glutamatergic synapse / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
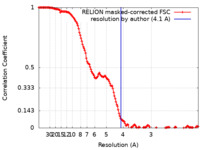
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Bogdanovic N / Tajima N | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis for kainate receptor activation by a partial agonist Authors: Bogdanovic N / Tajima N | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29926.map.gz emd_29926.map.gz | 25.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29926-v30.xml emd-29926-v30.xml emd-29926.xml emd-29926.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29926_fsc.xml emd_29926_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_29926.png emd_29926.png | 73.4 KB | ||
| Masks |  emd_29926_msk_1.map emd_29926_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29926.cif.gz emd-29926.cif.gz | 7 KB | ||
| Others |  emd_29926_additional_1.map.gz emd_29926_additional_1.map.gz emd_29926_half_map_1.map.gz emd_29926_half_map_1.map.gz emd_29926_half_map_2.map.gz emd_29926_half_map_2.map.gz | 6.6 MB 225.3 MB 225.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29926 http://ftp.pdbj.org/pub/emdb/structures/EMD-29926 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29926 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29926 | HTTPS FTP |
-Related structure data
| Related structure data |  8gc2MC  8gc3C  8gc4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29926.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29926.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Domoate-bound GluK2 kainate receptor in partially-open conformation 1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29926_msk_1.map emd_29926_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Additional Map
| File | emd_29926_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 1
| File | emd_29926_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_29926_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homotetrameric GluK2 bound with domoate state 1
| Entire | Name: Homotetrameric GluK2 bound with domoate state 1 |
|---|---|
| Components |
|
-Supramolecule #1: Homotetrameric GluK2 bound with domoate state 1
| Supramolecule | Name: Homotetrameric GluK2 bound with domoate state 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Tetramer isolated and dyalised extensively. The domoate was applied prior to grid freezing. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glutamate receptor ionotropic, kainate 2
| Macromolecule | Name: Glutamate receptor ionotropic, kainate 2 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 98.883469 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HVLRFGGIFE YVESGPMGAE ELAFRFAVNT INRNRTLLPN TTLTYDTQKI NLYDSFEASK KACDQLSLGV AAIFGPSHSS SANAVQSIC NALGVPHIQT RWKHQVSDNK DSFYVSLYPD FSSLSRAILD LVQFFKWKTV TVVYDDSTGL IRLQELIKAP S RYNLRLKI ...String: HVLRFGGIFE YVESGPMGAE ELAFRFAVNT INRNRTLLPN TTLTYDTQKI NLYDSFEASK KACDQLSLGV AAIFGPSHSS SANAVQSIC NALGVPHIQT RWKHQVSDNK DSFYVSLYPD FSSLSRAILD LVQFFKWKTV TVVYDDSTGL IRLQELIKAP S RYNLRLKI RQLPADTKDA KPLLKEMKRG KEFHVIFDCS HEMAAGILKQ ALAMGMMTEY YHYIFTTLDL FALDVEPYRY SG VNMTGFR ILNTENTQVS SIIEKWSMER LQAPPKPDSG LLDGFMTTDA ALMYDAVHVV SVAVQQFPQM TVSSLQCNRH KPW RFGTRF MSLIKEAHWE GLTGRITFNK TNGLRTDFDL DVISLKEEGL EKIGTWDPAS GLNMTESQKG KPANITDSLS NRSL IVTTI LEEPYVLFKK SDKPLYGNDR FEGYCIDLLR ELSTILGFTY EIRLVEDGKY GAQDDVNGQW NGMVRELIDH KADLA VAPL AITYVREKVI DFSKPFMTLG ISILYRKPNG TNPGVFSFLN PLSPDIWMYV LLAYLGVSVV LFVIARFSPY EWYNPH PSN PDSDVVENNF TLLNSFWFGV GALMQQGSEL MPKALSTRIV GGIWWFFTLI IISSYTANLA AFLTVERMES PIDSADD LA KQTKIEYGAV EDGATMTFFK KSKISTYDKM WAFMSSRRQS VLVKSNEEGI QRVLTSDYAF LMESTTIEFV TQRNCNLT Q IGGLIDSKGY GVGTPMGSPY RDKITIAILQ LQEEGKLHMM KEKWWRGNGC PEEESKEASA LGVQNIGGIF IVLAAGLVL SVFVAVGEFL YKSKKNAQLE KRSFCSAMVE ELRMSLKCQR RLKHKPQAPV IVKTEEVINM HTFNDRRLPG KETMA UniProtKB: Glutamate receptor ionotropic, kainate 2 |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 8 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIE...
| Macromolecule | Name: (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID type: ligand / ID: 7 / Number of copies: 3 / Formula: DOQ |
|---|---|
| Molecular weight | Theoretical: 311.33 Da |
| Chemical component information |  ChemComp-DOQ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Concentration: 150.0 mmol / Component - Formula: NaCl / Component - Name: sodium chloride / Details: 50 mM Tris/HCl 150 mM NaCl 1mM DDM pH 8.0 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
| Details | The sample was isolated from SEC and concentrated. Monodispersity of the final sample determined by the analytical HPLC, S200i column. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 12486 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 30 / Target criteria: 0.85 |
|---|---|
| Output model |  PDB-8gc2: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)