[English] 日本語
 Yorodumi
Yorodumi- EMDB-29879: Exploiting Activation and Inactivation Mechanisms in Type I-C CRI... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / type I-C / Cascade / Anti-CRISPR / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / hydrolase activity / RNA binding Similarity search - Function | |||||||||
| Biological species |  Neisseria lactamica (bacteria) Neisseria lactamica (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Hu C / Nam KH / Ke A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications. Authors: Chunyi Hu / Mason T Myers / Xufei Zhou / Zhonggang Hou / Macy L Lozen / Ki Hyun Nam / Yan Zhang / Ailong Ke /    Abstract: Type I CRISPR-Cas systems utilize the RNA-guided Cascade complex to identify matching DNA targets and the nuclease-helicase Cas3 to degrade them. Among the seven subtypes, type I-C is compact in size ...Type I CRISPR-Cas systems utilize the RNA-guided Cascade complex to identify matching DNA targets and the nuclease-helicase Cas3 to degrade them. Among the seven subtypes, type I-C is compact in size and highly active in creating large-sized genome deletions in human cells. Here, we use four cryoelectron microscopy snapshots to define its RNA-guided DNA binding and cleavage mechanisms in high resolution. The non-target DNA strand (NTS) is accommodated by I-C Cascade in a continuous binding groove along the juxtaposed Cas11 subunits. Binding of Cas3 further traps a flexible bulge in NTS, enabling NTS nicking. We identified two anti-CRISPR proteins AcrIC8 and AcrIC9 that strongly inhibit Neisseria lactamica I-C function. Structural analysis showed that AcrIC8 inhibits PAM recognition through allosteric inhibition, whereas AcrIC9 achieves so through direct competition. Both Acrs potently inhibit I-C-mediated genome editing and transcriptional modulation in human cells, providing the first off-switches for type I CRISPR eukaryotic genome engineering. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29879.map.gz emd_29879.map.gz | 52.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29879-v30.xml emd-29879-v30.xml emd-29879.xml emd-29879.xml | 33.7 KB 33.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29879.png emd_29879.png | 135.4 KB | ||
| Filedesc metadata |  emd-29879.cif.gz emd-29879.cif.gz | 8.3 KB | ||
| Others |  emd_29879_additional_1.map.gz emd_29879_additional_1.map.gz emd_29879_additional_2.map.gz emd_29879_additional_2.map.gz emd_29879_half_map_1.map.gz emd_29879_half_map_1.map.gz emd_29879_half_map_2.map.gz emd_29879_half_map_2.map.gz | 97.2 MB 97.2 MB 52.2 MB 52.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29879 http://ftp.pdbj.org/pub/emdb/structures/EMD-29879 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29879 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29879 | HTTPS FTP |
-Related structure data
| Related structure data |  8g9uMC  8g9sC  8g9tC  8gafC  8gamC  8ganC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29879.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29879.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.271 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #2
| File | emd_29879_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
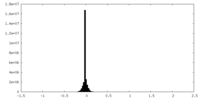
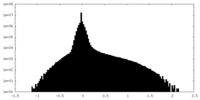
| Density Histograms |
-Additional map: #1
| File | emd_29879_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
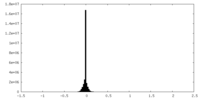
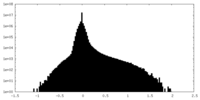
| Density Histograms |
-Half map: #2
| File | emd_29879_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29879_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Ternary complex of Cas3 with dsDNA bound type I-C Cascade at R-lo...
+Supramolecule #1: Ternary complex of Cas3 with dsDNA bound type I-C Cascade at R-lo...
+Macromolecule #1: Cas3
+Macromolecule #2: CRISPR-associated protein, Csd2 family
+Macromolecule #3: Phage associated protein
+Macromolecule #4: CRISPR-associated protein, Csd1 family
+Macromolecule #7: pre-crRNA processing endonuclease
+Macromolecule #5: crRNA (43-MER)
+Macromolecule #6: Traget strand DNA (53-MER)
+Macromolecule #8: proximal nontarget strand DNA (5'-D(P*AP*TP*GP*AP*AP*CP*TP*TP*CP*...
+Macromolecule #9: Distal nontarget DNA (5'-D(P*TP*TP*AP*TP*AP*TP*TP*AP*AP*TP*AP*TP*...
+Macromolecule #10: MAGNESIUM ION
+Macromolecule #11: PHOSPHATE ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 150.0 mM / Component - Formula: NaCl / Component - Name: sodium chloride / Details: 25mM Tris pH 7.5, 150mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 70.0 K / Max: 100.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1798 / Number real images: 1798 / Average exposure time: 2.5 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 67000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8g9u: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)