+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human cardiac myosin filament | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | cardiac / myosin / filament / complex / CONTRACTILE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmyosin II heavy chain binding / C zone / regulation of muscle filament sliding / muscle cell fate specification / striated muscle myosin thick filament / regulation of slow-twitch skeletal muscle fiber contraction / regulation of the force of skeletal muscle contraction / sarcomerogenesis / titin-telethonin complex / structural molecule activity conferring elasticity ...myosin II heavy chain binding / C zone / regulation of muscle filament sliding / muscle cell fate specification / striated muscle myosin thick filament / regulation of slow-twitch skeletal muscle fiber contraction / regulation of the force of skeletal muscle contraction / sarcomerogenesis / titin-telethonin complex / structural molecule activity conferring elasticity / skeletal muscle myosin thick filament assembly / telethonin binding / regulation of striated muscle contraction / A band / cardiac myofibril / detection of muscle stretch / muscle myosin complex / : / muscle alpha-actinin binding / cardiac myofibril assembly / regulation of the force of heart contraction / transition between fast and slow fiber / myosin filament / cardiac muscle hypertrophy / cardiac muscle tissue morphogenesis / mitotic chromosome condensation / protein kinase regulator activity / adult heart development / Striated Muscle Contraction / muscle filament sliding / cardiac muscle hypertrophy in response to stress / regulation of cardiac muscle cell contraction / myosin complex / M band / actinin binding / myosin II complex / cardiac muscle cell development / I band / structural constituent of muscle / sarcomere organization / ventricular cardiac muscle tissue morphogenesis / microfilament motor activity / myosin heavy chain binding / heart contraction / myosin binding / myofibril / positive regulation of the force of heart contraction / ATPase activator activity / cytoskeletal motor activity / striated muscle thin filament / skeletal muscle thin filament assembly / actin monomer binding / heart morphogenesis / striated muscle contraction / ATP metabolic process / skeletal muscle tissue development / skeletal muscle contraction / cardiac muscle contraction / stress fiber / titin binding / regulation of heart rate / muscle contraction / sarcomere / condensed nuclear chromosome / positive regulation of protein secretion / post-embryonic development / response to calcium ion / negative regulation of cell growth / Z disc / actin filament binding / Platelet degranulation / heart development / actin binding / protease binding / protein tyrosine kinase activity / cytoskeleton / calmodulin binding / protein kinase activity / non-specific serine/threonine protein kinase / cell adhesion / protein serine kinase activity / protein serine/threonine kinase activity / calcium ion binding / positive regulation of gene expression / protein kinase binding / enzyme binding / protein homodimerization activity / extracellular exosome / extracellular region / ATP binding / metal ion binding / identical protein binding / cytoplasm / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
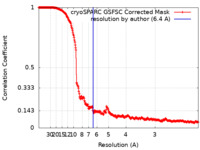
| Method | single particle reconstruction / cryo EM / Resolution: 6.4 Å | |||||||||||||||
 Authors Authors | Dutta D / Nguyen V / Padron R / Craig R | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Cryo-EM structure of the human cardiac myosin filament. Authors: Debabrata Dutta / Vu Nguyen / Kenneth S Campbell / Raúl Padrón / Roger Craig /  Abstract: Pumping of the heart is powered by filaments of the motor protein myosin that pull on actin filaments to generate cardiac contraction. In addition to myosin, the filaments contain cardiac myosin- ...Pumping of the heart is powered by filaments of the motor protein myosin that pull on actin filaments to generate cardiac contraction. In addition to myosin, the filaments contain cardiac myosin-binding protein C (cMyBP-C), which modulates contractility in response to physiological stimuli, and titin, which functions as a scaffold for filament assembly. Myosin, cMyBP-C and titin are all subject to mutation, which can lead to heart failure. Despite the central importance of cardiac myosin filaments to life, their molecular structure has remained a mystery for 60 years. Here we solve the structure of the main (cMyBP-C-containing) region of the human cardiac filament using cryo-electron microscopy. The reconstruction reveals the architecture of titin and cMyBP-C and shows how myosin's motor domains (heads) form three different types of motif (providing functional flexibility), which interact with each other and with titin and cMyBP-C to dictate filament architecture and function. The packing of myosin tails in the filament backbone is also resolved. The structure suggests how cMyBP-C helps to generate the cardiac super-relaxed state; how titin and cMyBP-C may contribute to length-dependent activation; and how mutations in myosin and cMyBP-C might disturb interactions, causing disease. The reconstruction resolves past uncertainties and integrates previous data on cardiac muscle structure and function. It provides a new paradigm for interpreting structural, physiological and clinical observations, and for the design of potential therapeutic drugs. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29722.map.gz emd_29722.map.gz | 1.7 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29722-v30.xml emd-29722-v30.xml emd-29722.xml emd-29722.xml | 33.2 KB 33.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29722_fsc.xml emd_29722_fsc.xml | 26.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_29722.png emd_29722.png | 119.5 KB | ||
| Filedesc metadata |  emd-29722.cif.gz emd-29722.cif.gz | 8.8 KB | ||
| Others |  emd_29722_additional_1.map.gz emd_29722_additional_1.map.gz emd_29722_half_map_1.map.gz emd_29722_half_map_1.map.gz emd_29722_half_map_2.map.gz emd_29722_half_map_2.map.gz | 1.8 GB 1.8 GB 1.8 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29722 http://ftp.pdbj.org/pub/emdb/structures/EMD-29722 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29722 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29722 | HTTPS FTP |
-Related structure data
| Related structure data |  8g4lMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29722.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29722.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0873 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_29722_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29722_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29722_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Myosin filaments isolated from human cardiac left ventricular muscle
| Entire | Name: Myosin filaments isolated from human cardiac left ventricular muscle |
|---|---|
| Components |
|
-Supramolecule #1: Myosin filaments isolated from human cardiac left ventricular muscle
| Supramolecule | Name: Myosin filaments isolated from human cardiac left ventricular muscle type: tissue / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Myosin-7
| Macromolecule | Name: Myosin-7 / type: protein_or_peptide / ID: 1 / Number of copies: 78 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: heart Homo sapiens (human) / Tissue: heart |
| Molecular weight | Theoretical: 223.445984 KDa |
| Sequence | String: MGDSEMAVFG AAAPYLRKSE KERLEAQTRP FDLKKDVFVP DDKQEFVKAK IVSREGGKVT AETEYGKTVT VKEDQVMQQN PPKFDKIED MAMLTFLHEP AVLYNLKDRY GSWMIYTYSG LFCVTVNPYK WLPVYTPEVV AAYRGKKRSE APPHIFSISD N AYQYMLTD ...String: MGDSEMAVFG AAAPYLRKSE KERLEAQTRP FDLKKDVFVP DDKQEFVKAK IVSREGGKVT AETEYGKTVT VKEDQVMQQN PPKFDKIED MAMLTFLHEP AVLYNLKDRY GSWMIYTYSG LFCVTVNPYK WLPVYTPEVV AAYRGKKRSE APPHIFSISD N AYQYMLTD RENQSILITG ESGAGKTVNT KRVIQYFAVI AAIGDRSKKD QSPGKGTLED QIIQANPALE AFGNAKTVRN DN SSRFGKF IRIHFGATGK LASADIETYL LEKSRVIFQL KAERDYHIFY QILSNKKPEL LDMLLITNNP YDYAFISQGE TTV ASIDDA EELMATDNAF DVLGFTSEEK NSMYKLTGAI MHFGNMKFKL KQREEQAEPD GTEEADKSAY LMGLNSADLL KGLC HPRVK VGNEYVTKGQ NVQQVIYATG ALAKAVYERM FNWMVTRINA TLETKQPRQY FIGVLDIAGF EIFDFNSFEQ LCINF TNEK LQQFFNHHMF VLEQEEYKKE GIEWTFIDFG MDLQACIDLI EKPMGIMSIL EEECMFPKAT DMTFKAKLFD NHLGKS ANF QKPRNIKGKP EAHFSLIHYA GIVDYNIIGW LQKNKDPLNE TVVGLYQKSS LKLLSTLFAN YAGADAPIEK GKGKAKK GS SFQTVSALHR ENLNKLMTNL RSTHPHFVRC IIPNETKSPG VMDNPLVMHQ LRCNGVLEGI RICRKGFPNR ILYGDFRQ R YRILNPAAIP EGQFIDSRKG AEKLLSSLDI DHNQYKFGHT KVFFKAGLLG LLEEMRDERL SRIITRIQAQ SRGVLARME YKKLLERRDS LLVIQWNIRA FMGVKNWPWM KLYFKIKPLL KSAEREKEMA SMKEEFTRLK EALEKSEARR KELEEKMVSL LQEKNDLQL QVQAEQDNLA DAEERCDQLI KNKIQLEAKV KEMNERLEDE EEMNAELTAK KRKLEDECSE LKRDIDDLEL T LAKVEKEK HATENKVKNL TEEMAGLDEI IAKLTKEKKA LQEAHQQALD DLQAEEDKVN TLTKAKVKLE QQVDDLEGSL EQ EKKVRMD LERAKRKLEG DLKLTQESIM DLENDKQQLD ERLKKKDFEL NALNARIEDE QALGSQLQKK LKELQARIEE LEE ELEAER TARAKVEKLR SDLSRELEEI SERLEEAGGA TSVQIEMNKK REAEFQKMRR DLEEATLQHE ATAAALRKKH ADSV AELGE QIDNLQRVKQ KLEKEKSEFK LELDDVTSNM EQIIKAKANL EKMCRTLEDQ MNEHRSKAEE TQRSVNDLTS QRAKL QTEN GELSRQLDEK EALISQLTRG KLTYTQQLED LKRQLEEEVK AKNALAHALQ SARHDCDLLR EQYEEETEAK AELQRV LSK ANSEVAQWRT KYETDAIQRT EELEEAKKKL AQRLQEAEEA VEAVNAKCSS LEKTKHRLQN EIEDLMVDVE RSNAAAA AL DKKQRNFDKI LAEWKQKYEE SQSELESSQK EARSLSTELF KLKNAYEESL EHLETFKREN KNLQEEISDL TEQLGSSG K TIHELEKVRK QLEAEKMELQ SALEEAEASL EHEEGKILRA QLEFNQIKAE IERKLAEKDE EMEQAKRNHL RVVDSLQTS LDAETRSRNE ALRVKKKMEG DLNEMEIQLS HANRMAAEAQ KQVKSLQSLL KDTQIQLDDA VRANDDLKEN IAIVERRNNL LQAELEELR AVVEQTERSR KLAEQELIET SERVQLLHSQ NTSLINQKKK MDADLSQLQT EVEEAVQECR NAEEKAKKAI T DAAMMAEE LKKEQDTSAH LERMKKNMEQ TIKDLQHRLD EAEQIALKGG KKQLQKLEAR VRELENELEA EQKRNAESVK GM RKSERRI KELTYQTEED RKNLLRLQDL VDKLQLKVKA YKRQAEEAEE QANTNLSKFR KVQHELDEAE ERADIAESQV NKL RAKSRD IGTKGLNEE UniProtKB: Myosin-7 |
-Macromolecule #2: Myosin light chain 3
| Macromolecule | Name: Myosin light chain 3 / type: protein_or_peptide / ID: 2 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: heart Homo sapiens (human) / Tissue: heart |
| Molecular weight | Theoretical: 21.962068 KDa |
| Sequence | String: MAPKKPEPKK DDAKAAPKAA PAPAPPPEPE RPKEVEFDAS KIKIEFTPEQ IEEFKEAFML FDRTPKCEMK ITYGQCGDVL RALGQNPTQ AEVLRVLGKP RQEELNTKMM DFETFLPMLQ HISKNKDTGT YEDFVEGLRV FDKEGNGTVM GAELRHVLAT L GERLTEDE ...String: MAPKKPEPKK DDAKAAPKAA PAPAPPPEPE RPKEVEFDAS KIKIEFTPEQ IEEFKEAFML FDRTPKCEMK ITYGQCGDVL RALGQNPTQ AEVLRVLGKP RQEELNTKMM DFETFLPMLQ HISKNKDTGT YEDFVEGLRV FDKEGNGTVM GAELRHVLAT L GERLTEDE VEKLMAGQED SNGCINYEAF VKHIMSS UniProtKB: Myosin light chain 3 |
-Macromolecule #3: Myosin regulatory light chain 2, ventricular/cardiac muscle isoform
| Macromolecule | Name: Myosin regulatory light chain 2, ventricular/cardiac muscle isoform type: protein_or_peptide / ID: 3 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: heart Homo sapiens (human) / Tissue: heart |
| Molecular weight | Theoretical: 18.813273 KDa |
| Sequence | String: MAPKKAKKRA GGANSNVFSM FEQTQIQEFK EAFTIMDQNR DGFIDKNDLR DTFAALGRVN VKNEEIDEMI KEAPGPINFT VFLTMFGEK LKGADPEETI LNAFKVFDPE GKGVLKADYV REMLTTQAER FSKEEVDQMF AAFPPDVTGN LDYKNLVHII T HGEEKD UniProtKB: Myosin regulatory light chain 2, ventricular/cardiac muscle isoform |
-Macromolecule #4: Titin
| Macromolecule | Name: Titin / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: heart Homo sapiens (human) / Tissue: heart |
| Molecular weight | Theoretical: 119.771961 KDa |
| Sequence | String: PPEIELDADL RKVVTIRACC TLRLFVPIKG RPAPEVKWAR DHGESLDKAS IESTSSYTLL IVGNVNRFDS GKYILTVENS SGSKSAFVN VRVLDTPGPP QDLKVKEVTK TSVTLTWDPP LLDGGSKIKN YIVEKRESTR KAYSTVATNC HKTSWKVDQL Q EGCSYYFR ...String: PPEIELDADL RKVVTIRACC TLRLFVPIKG RPAPEVKWAR DHGESLDKAS IESTSSYTLL IVGNVNRFDS GKYILTVENS SGSKSAFVN VRVLDTPGPP QDLKVKEVTK TSVTLTWDPP LLDGGSKIKN YIVEKRESTR KAYSTVATNC HKTSWKVDQL Q EGCSYYFR VLAENEYGIG LPAETAESVK ASERPLPPGK ITLMDVTRNS VSLSWEKPEH DGGSRILGYI VEMQTKGSDK WA TCATVKV TEATITGLIQ GEEYSFRVSA QNEKGISDPR QLSVPVIAKD LVIPPAFKLL FNTFTVLAGE DLKVDVPFIG RPT PAVTWH KDNVPLKQTT RVNAESTENN SLLTIKDACR EDVGHYVVKL TNSAGEAIET LNVIVLDKPG PPTGPVKMDE VTAD SITLS WGPPKYDGGS SINNYIVEKR DTSTTTWQIV SATVARTTIK ACRLKTGCEY QFRIAAENRY GKSTYLNSEP TVAQY PFKV PGPPGTPVVT LSSRDSMEVQ WNEPISDGGS RVIGYHLERK ERNSILWVKL NKTPIPQTKF KTTGLEEGVE YEFRVS AEN IVGIGKPSKV SECYVARDPC DPPGRPEAII VTRNSVTLQW KKPTYDGGSK ITGYIVEKKE LPEGRWMKAS FTNIIDT HF EVTGLVEDHR YEFRVIARNA AGVFSEPSES TGAITARDEV DPPRISMDPK YKDTIVVHAG ESFKVDADIY GKPIPTIQ W IKGDQELSNT ARLEIKSTDF ATSLSVKDAV RVDSGNYILK AKNVAGERSV TVNVKVLDRP GPPEGPVVIS GVTAEKCTL AWKPPLQDGG SDIINYIVER RETSRLVWTV VDANVQTLSC KVTKLLEGNE YTFRIMAVNK YGVGEPLESE PVVAKNPFVV PDAPKAPEV TTVTKDSMIV VWERPASDGG SEILGYVLEK RDKEGIRWTR CHKRLIGELR LRVTGLIENH DYEFRVSAEN A AGLSEPSP PSAYQKACDP IYKPGPPNNP KVIDITRSSV FLSWSKPIYD GGCEIQGYIV EKCDVSVGEW TMCTPPTGIN KT NIEVEKL LEKHEYNFRI CAINKAGVGE HADVPGPIIV EEKLEAP UniProtKB: Titin |
-Macromolecule #5: Myosin-binding protein C, cardiac-type
| Macromolecule | Name: Myosin-binding protein C, cardiac-type / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: heart Homo sapiens (human) / Tissue: heart |
| Molecular weight | Theoretical: 140.947172 KDa |
| Sequence | String: MPEPGKKPVS AFSKKPRSVE VAAGSPAVFE AETERAGVKV RWQRGGSDIS ASNKYGLATE GTRHTLTVRE VGPADQGSYA VIAGSSKVK FDLKVIEAEK AEPMLAPAPA PAEATGAPGE APAPAAELGE SAPSPKGSSS AALNGPTPGA PDDPIGLFVM R PQDGEVTV ...String: MPEPGKKPVS AFSKKPRSVE VAAGSPAVFE AETERAGVKV RWQRGGSDIS ASNKYGLATE GTRHTLTVRE VGPADQGSYA VIAGSSKVK FDLKVIEAEK AEPMLAPAPA PAEATGAPGE APAPAAELGE SAPSPKGSSS AALNGPTPGA PDDPIGLFVM R PQDGEVTV GGSITFSARV AGASLLKPPV VKWFKGKWVD LSSKVGQHLQ LHDSYDRASK VYLFELHITD AQPAFTGSYR CE VSTKDKF DCSNFNLTVH EAMGTGDLDL LSAFRRTSLA GGGRRISDSH EDTGILDFSS LLKKRDSFRT PRDSKLEAPA EED VWEILR QAPPSEYERI AFQYGVTDLR GMLKRLKGMR RDEKKSTAFQ KKLEPAYQVS KGHKIRLTVE LADHDAEVKW LKNG QEIQM SGSKYIFESI GAKRTLTISQ CSLADDAAYQ CVVGGEKCST ELFVKEPPVL ITRPLEDQLV MVGQRVEFEC EVSEE GAQV KWLKDGVELT REETFKYRFK KDGQRHHLII NEAMLEDAGH YALCTSGGQA LAELIVQEKK LEVYQSIADL MVGAKD QAV FKCEVSDENV RGVWLKNGKE LVPDSRIKVS HIGRVHKLTI DDVTPADEAD YSFVPEGFAC NLSAKLHFME VKIDFVP RQ EPPKIHLDCP GRIPDTIVVV AGNKLRLDVP ISGDPAPTVI WQKAITQGNK APARPAPDAP EDTGDSDEWV FDKKLLCE T EGRVRVETTK DRSIFTVEGA EKEDEGVYTV TVKNPVGEDQ VNLTVKVIDV PDAPAAPKIS NVGEDSCTVQ WEPPAYDGG QPILGYILER KKKKSYRWMR LNFDLIQELS HEARRMIEGV VYEMRVYAVN AIGMSRPSPA SQPFMPIGPP SEPTHLAVED VSDTTVSLK WRPPERVGAG GLDGYSVEYC PEGCSEWVAA LQGLTEHTSI LVKDLPTGAR LLFRVRAHNM AGPGAPVTTT E PVTVQEIL QRPRLQLPRH LRQTIQKKVG EPVNLLIPFQ GKPRPQVTWT KEGQPLAGEE VSIRNSPTDT ILFIRAARRV HS GTYQVTV RIENMEDKAT LVLQVVDKPS PPQDLRVTDA WGLNVALEWK PPQDVGNTEL WGYTVQKADK KTMEWFTVLE HYR RTHCVV PELIIGNGYY FRVFSQNMVG FSDRAATTKE PVFIPRPGIT YEPPNYKALD FSEAPSFTQP LVNRSVIAGY TAML CCAVR GSPKPKISWF KNGLDLGEDA RFRMFSKQGV LTLEIRKPCP FDGGIYVCRA TNLQGEARCE CRLEVRVPQ UniProtKB: Myosin-binding protein C, cardiac-type |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 61.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8g4l: |
-Atomic model buiding 2
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-8g4l: |
-Atomic model buiding 3
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-8g4l: |
-Atomic model buiding 4
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-8g4l: |
-Atomic model buiding 5
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-8g4l: |
-Atomic model buiding 6
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-8g4l: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)