[English] 日本語
 Yorodumi
Yorodumi- EMDB-29646: Acto-HMM complex in ADP-state. Chicken smooth muscle HMM and chic... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acto-HMM complex in ADP-state. Chicken smooth muscle HMM and chicken pectoralis actin | ||||||||||||||||||||||||||||||
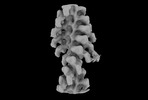
 Map data Map data | Two-headed attachment - Post-classification - filtered map | ||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | Actin / Myosin / Smooth muscle / Cryo-EM / Cryo-ET / Heavy meromyosin / MOTOR PROTEIN | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationRHO GTPases activate PAKs / myosin II filament / Smooth Muscle Contraction / elastic fiber assembly / myofibril assembly / skeletal muscle myosin thick filament assembly / myosin light chain binding / myosin II binding / Striated Muscle Contraction / muscle myosin complex ...RHO GTPases activate PAKs / myosin II filament / Smooth Muscle Contraction / elastic fiber assembly / myofibril assembly / skeletal muscle myosin thick filament assembly / myosin light chain binding / myosin II binding / Striated Muscle Contraction / muscle myosin complex / actomyosin / myosin filament / actomyosin structure organization / myosin complex / myosin II complex / cardiac muscle cell development / structural constituent of muscle / myosin heavy chain binding / microfilament motor activity / myofibril / cytoskeletal motor activity / striated muscle thin filament / skeletal muscle thin filament assembly / smooth muscle contraction / skeletal muscle fiber development / stress fiber / actin filament / ADP binding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / actin filament binding / actin cytoskeleton / actin binding / calmodulin binding / hydrolase activity / calcium ion binding / magnesium ion binding / ATP binding / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 19.0 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Hojjatian A / Liu J / Taylor DW / Daneshparvar N / Trybus KM / Taylor KA | ||||||||||||||||||||||||||||||
| Funding support |  United States, 9 items United States, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2023 Journal: J Struct Biol / Year: 2023Title: Double-headed binding of myosin II to F-actin shows the effect of strain on head structure. Authors: Alimohammad Hojjatian / Dianne W Taylor / Nadia Daneshparvar / Patricia M Fagnant / Kathleen M Trybus / Kenneth A Taylor /  Abstract: Force production in muscle is achieved through the interaction of myosin and actin. Strong binding states in active muscle are associated with Mg·ADP bound to the active site; release of Mg·ADP ...Force production in muscle is achieved through the interaction of myosin and actin. Strong binding states in active muscle are associated with Mg·ADP bound to the active site; release of Mg·ADP allows rebinding of ATP and dissociation from actin. Thus, Mg·ADP binding is positioned for adaptation as a force sensor. Mechanical loads on the lever arm can affect the ability of myosin to release Mg·ADP but exactly how this is done is poorly defined. Here we use F-actin decorated with double-headed smooth muscle myosin fragments in the presence of Mg·ADP to visualize the effect of internally supplied tension on the paired lever arms using cryoEM. The interaction of the paired heads with two adjacent actin subunits is predicted to place one lever arm under positive and the other under negative strain. The converter domain is believed to be the most flexible domain within myosin head. Our results, instead, point to the segment of heavy chain between the essential and regulatory light chains as the location of the largest structural change. Moreover, our results suggest no large changes in the myosin coiled coil tail as the locus of strain relief when both heads bind F-actin. The method would be adaptable to double-headed members of the myosin family. We anticipate that the study of actin-myosin interaction using double-headed fragments enables visualization of domains that are typically noisy in decoration with single-headed fragments. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29646.map.gz emd_29646.map.gz | 16.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29646-v30.xml emd-29646-v30.xml emd-29646.xml emd-29646.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29646_fsc.xml emd_29646_fsc.xml | 7.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_29646.png emd_29646.png | 28.7 KB | ||
| Others |  emd_29646_additional_1.map.gz emd_29646_additional_1.map.gz emd_29646_half_map_1.map.gz emd_29646_half_map_1.map.gz emd_29646_half_map_2.map.gz emd_29646_half_map_2.map.gz | 27.3 MB 27.4 MB 27.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29646 http://ftp.pdbj.org/pub/emdb/structures/EMD-29646 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29646 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29646 | HTTPS FTP |
-Related structure data
| Related structure data |  8g06  8syfMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29646.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29646.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Two-headed attachment - Post-classification - filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.56 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Acto-HMM - Pre-classification - filtered map
| File | emd_29646_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Acto-HMM - Pre-classification - filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Two-headed attachment - Post-classification - unfiltered half map 1
| File | emd_29646_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Two-headed attachment - Post-classification - unfiltered half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Two-headed attachment - Post-classification - unfiltered half map 2
| File | emd_29646_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Two-headed attachment - Post-classification - unfiltered half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Acto-HMM complex - chicken smooth muscle heavy meromyosin in comp...
| Entire | Name: Acto-HMM complex - chicken smooth muscle heavy meromyosin in complex with chicken pectoralis muscle actin filament |
|---|---|
| Components |
|
-Supramolecule #1: Acto-HMM complex - chicken smooth muscle heavy meromyosin in comp...
| Supramolecule | Name: Acto-HMM complex - chicken smooth muscle heavy meromyosin in complex with chicken pectoralis muscle actin filament type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Myosin regulatory light chain 2, smooth muscle major isoform
| Macromolecule | Name: Myosin regulatory light chain 2, smooth muscle major isoform type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.583412 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: FDQSQIQEFK EAFNMIDQNR DGFIDKEDLH DMLASMGKNP TDEYLEGMMS EAPGPINFTM FLTMFGEKLN GTDPEDVIRN AFACFDEEA SGFIHEDHLR ELLTTMGDRF TDEEVDEMYR EAPIDKKGNF NYVEFTRILK HGAK UniProtKB: Myosin regulatory light chain 2, smooth muscle major isoform |
-Macromolecule #2: Myosin-11
| Macromolecule | Name: Myosin-11 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 97.534883 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: SQKPLSDDEK FLFVDKNFVN NPLAQADWSA KKLVWVPSEK HGFEAASIKE EKGDEVTVEL QENGKKVTLS KDDIQKMNPP KFSKVEDMA ELTCLNEASV LHNLRERYFS GLIYTYSGLF CVVINPYKQL PIYSEKIIDM YKGKKRHEMP PHIYAIADTA Y RSMLQDRE ...String: SQKPLSDDEK FLFVDKNFVN NPLAQADWSA KKLVWVPSEK HGFEAASIKE EKGDEVTVEL QENGKKVTLS KDDIQKMNPP KFSKVEDMA ELTCLNEASV LHNLRERYFS GLIYTYSGLF CVVINPYKQL PIYSEKIIDM YKGKKRHEMP PHIYAIADTA Y RSMLQDRE DQSILCTGES GAGKTENTKK VIQYLAVVAS SHKGKKDTSI TQGPSFSYGE LEKQLLQANP ILEAFGNAKT VK NDNSSRF GKFIRINFDV TGYIVGANIE TYLLEKSRAI RQAKDERTFH IFYYLIAGAS EQMRNDLLLE GFNNYTFLSN GHV PIPAQQ DDEMFQETLE AMTIMGFTEE EQTSILRVVS SVLQLGNIVF KKERNTDQAS MPDNTAAQKV CHLMGINVTD FTRS ILTPR IKVGRDVVQK AQTKEQADFA IEALAKAKFE RLFRWILTRV NKALDKTKRQ GASFLGILDI AGFEIFEINS FEQLC INYT NEKLQQLFNH TMFILEQEEY QREGIEWNFI DFGLDLQPCI ELIERPTNPP GVLALLDEEC WFPKATDTSF VEKLIQ EQG NHAKFQKSKQ LKDKTEFCIL HYAGKVTYNA SAWLTKNMDP LNDNVTSLLN QSSDKFVADL WKDVDRIVGL DQMAKMT ES SLPSASKTKK GMFRTVGQLY KEQLTKLMTT LRNTNPNFVR CIIPNHEKRA GKLDAHLVLE QLRCNGVLEG IRICRQGF P NRIVFQEFRQ RYEILAANAI PKGFMDGKQA CILMIKALEL DPNLYRIGQS KIFFRTGVLA HLEEERDLKI TDVIIAFQA QCRGYLARKA FAKRQQQLTA MKVIQRNCAA YLKLRNWQWW RLFTKVKPLL UniProtKB: Myosin-11 |
-Macromolecule #3: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.862613 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: DEDETTALVC DNGSGLVKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIITNWD DMEKIWHHT FYNELRVAPE EHPTLLTEAP LNPKANREKM TQIMFETFNV PAMYVAIQAV LSLYASGRTT GIVLDSGDGV T HNVPIYEG ...String: DEDETTALVC DNGSGLVKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIITNWD DMEKIWHHT FYNELRVAPE EHPTLLTEAP LNPKANREKM TQIMFETFNV PAMYVAIQAV LSLYASGRTT GIVLDSGDGV T HNVPIYEG YALPHAIMRL DLAGRDLTDY LMKILTERGY SFVTTAEREI VRDIKEKLCY VALDFENEMA TAASSSSLEK SY ELPDGQV ITIGNERFRC PETLFQPSFI GMESAGIHET TYNSIMKCDI DIRKDLYANN VMSGGTTMYP GIADRMQKEI TAL APSTMK IKIIAPPERK YSVWIGGSIL ASLSTFQQMW ITKQEYDEAG PSIVHRKCF UniProtKB: Actin, alpha skeletal muscle |
-Macromolecule #4: Myosin light polypeptide 6
| Macromolecule | Name: Myosin light polypeptide 6 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.639715 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: FSEEQTAEFK EAFQLFDRTG DGKILYSQCG DVMRALGQNP TNAEVMKVLG NPKSDEMNLK TLNFEQFLPM MQTIAKNKDQ GCFEDYVEG LRVFDKEGNG TVMGAEIRHV LVTLGEKMTE EEVEQLVAGH EDSNGCINYE ELVRMVLSG UniProtKB: Myosin light polypeptide 6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-64 (8k x 8k) / Detector mode: INTEGRATING / Average electron dose: 29.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 20.0 µm / Nominal defocus min: 0.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































