+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human TMEM175-LAMP1 full-length complex | |||||||||
 Map data Map data | TMEM175-LAMP1 full-length complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlysosomal lumen pH elevation / phagosome-lysosome fusion / regulation of lysosomal lumen pH / potassium ion leak channel activity / proton channel activity / arachidonate binding / potassium channel activity / potassium ion transmembrane transport / proton transmembrane transport / neuron cellular homeostasis ...lysosomal lumen pH elevation / phagosome-lysosome fusion / regulation of lysosomal lumen pH / potassium ion leak channel activity / proton channel activity / arachidonate binding / potassium channel activity / potassium ion transmembrane transport / proton transmembrane transport / neuron cellular homeostasis / large ribosomal subunit / endosome membrane / lysosome / endosome / structural constituent of ribosome / translation / lysosomal membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
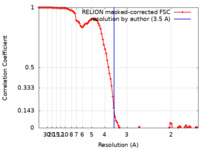
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Zhang JY / Zeng WZ / Han Y / Jiang YX | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel. Authors: Jiyuan Zhang / Weizhong Zeng / Yan Han / Wan-Ru Lee / Jen Liou / Youxing Jiang /  Abstract: Maintaining a highly acidic lysosomal pH is central to cellular physiology. Here, we use functional proteomics, single-particle cryo-EM, electrophysiology, and in vivo imaging to unravel a key ...Maintaining a highly acidic lysosomal pH is central to cellular physiology. Here, we use functional proteomics, single-particle cryo-EM, electrophysiology, and in vivo imaging to unravel a key biological function of human lysosome-associated membrane proteins (LAMP-1 and LAMP-2) in regulating lysosomal pH homeostasis. Despite being widely used as a lysosomal marker, the physiological functions of the LAMP proteins have long been overlooked. We show that LAMP-1 and LAMP-2 directly interact with and inhibit the activity of the lysosomal cation channel TMEM175, a key player in lysosomal pH homeostasis implicated in Parkinson's disease. This LAMP inhibition mitigates the proton conduction of TMEM175 and facilitates lysosomal acidification to a lower pH environment crucial for optimal hydrolase activity. Disrupting the LAMP-TMEM175 interaction alkalinizes the lysosomal pH and compromises the lysosomal hydrolytic function. In light of the ever-increasing importance of lysosomes to cellular physiology and diseases, our data have widespread implications for lysosomal biology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29553.map.gz emd_29553.map.gz | 85.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29553-v30.xml emd-29553-v30.xml emd-29553.xml emd-29553.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29553_fsc.xml emd_29553_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_29553.png emd_29553.png | 56.9 KB | ||
| Filedesc metadata |  emd-29553.cif.gz emd-29553.cif.gz | 5.6 KB | ||
| Others |  emd_29553_half_map_1.map.gz emd_29553_half_map_1.map.gz emd_29553_half_map_2.map.gz emd_29553_half_map_2.map.gz | 69.9 MB 69.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29553 http://ftp.pdbj.org/pub/emdb/structures/EMD-29553 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29553 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29553 | HTTPS FTP |
-Validation report
| Summary document |  emd_29553_validation.pdf.gz emd_29553_validation.pdf.gz | 789 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29553_full_validation.pdf.gz emd_29553_full_validation.pdf.gz | 788.6 KB | Display | |
| Data in XML |  emd_29553_validation.xml.gz emd_29553_validation.xml.gz | 17.3 KB | Display | |
| Data in CIF |  emd_29553_validation.cif.gz emd_29553_validation.cif.gz | 22.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29553 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29553 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29553 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29553 | HTTPS FTP |
-Related structure data
| Related structure data |  8fy5MC  8fyfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29553.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29553.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TMEM175-LAMP1 full-length complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29553_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29553_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TMEM175-LAMP1 full-length
| Entire | Name: TMEM175-LAMP1 full-length |
|---|---|
| Components |
|
-Supramolecule #1: TMEM175-LAMP1 full-length
| Supramolecule | Name: TMEM175-LAMP1 full-length / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Endosomal/lysosomal potassium channel TMEM175
| Macromolecule | Name: Endosomal/lysosomal potassium channel TMEM175 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.667219 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSQPRTPEQA LDTPGDCPPG RRDEDAGEGI QCSQRMLSFS DALLSIIATV MILPVTHTEI SPEQQFDRSV QRLLATRIAV YLMTFLIVT VAWAAHTRLF QVVGKTDDTL ALLNLACMMT ITFLPYTFSL MVTFPDVPLG IFLFCVCVIA IGVVQALIVG Y AFHFPHLL ...String: MSQPRTPEQA LDTPGDCPPG RRDEDAGEGI QCSQRMLSFS DALLSIIATV MILPVTHTEI SPEQQFDRSV QRLLATRIAV YLMTFLIVT VAWAAHTRLF QVVGKTDDTL ALLNLACMMT ITFLPYTFSL MVTFPDVPLG IFLFCVCVIA IGVVQALIVG Y AFHFPHLL SPQIQRSAHR ALYRRHVLGI VLQGPALCFA AAIFSLFFVP LSYLLMVTVI LLPYVSKVTG WCRDRLLGHR EP SAHPVEV FSFDLHEPLS KERVEAFSDG VYAIVATLLI LDICEDNVPD PKDVKERFSG SLVAALSATG PRFLAYFGSF ATV GLLWFA HHSLFLHVRK ATRAMGLLNT LSLAFVGGLP LAYQQTSAFA RQPRDELERV RVSCTIIFLA SIFQLAMWTT ALLH QAETL QPSVWFGGRE HVLMFAKLAL YPCASLLAFA STCLLSRFSV GIFHLMQIAV PCAFLLLRLL VGLALATLRV LRGLA RPEH PPPAPTGQDD PQSQLLPAPC UniProtKB: Endosomal/lysosomal proton channel TMEM175 |
-Macromolecule #2: Lysosome-associated membrane glycoprotein 1
| Macromolecule | Name: Lysosome-associated membrane glycoprotein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.928215 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAPGSARRP LLLLLLLLLL GLMHCASAAM FMVKNGNGTA CIMANFSAAF SVNYDTKSGP KNMTFDLPSD ATVVLNRSSC GKENTSDPS LVIAFGRGHT LTLNFTRNAT RYSVQLMSFV YNLSDTHLFP NASSKEIKTV ESITDIRADI DKKYRCVSGT Q VHMNNVTV ...String: MAAPGSARRP LLLLLLLLLL GLMHCASAAM FMVKNGNGTA CIMANFSAAF SVNYDTKSGP KNMTFDLPSD ATVVLNRSSC GKENTSDPS LVIAFGRGHT LTLNFTRNAT RYSVQLMSFV YNLSDTHLFP NASSKEIKTV ESITDIRADI DKKYRCVSGT Q VHMNNVTV TLHDATIQAY LSNSSFSRGE TRCEQDRPSP TTAPPAPPSP SPSPVPKSPS VDKYNVSGTN GTCLLASMGL QL NLTYERK DNTTVTRLLN INPNKTSASG SCGAHLVTLE LHSEGTTVLL FQFGMNASSS RFFLQGIQLN TILPDARDPA FKA ANGSLR ALQATVGNSY KCNAEEHVRV TKAFSVNIFK VWVQAFKVEG GQFGSVEECL LDENSMLIPI AVGGALAGLV LIVL IAYLV GRKRSHAGYQ TI UniProtKB: Ribosomal protein L22p/L17e |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8fy5: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)