[English] 日本語
 Yorodumi
Yorodumi- EMDB-29524: Human M4 muscarinic acetylcholine receptor complex with Gi1 and x... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human M4 muscarinic acetylcholine receptor complex with Gi1 and xanomeline | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | 7 transmembrane receptor / SIGNALING PROTEIN-IMMUNE SYSTEM complex / MEMBRANE PROTEIN / MEMBRANE PROTEIN-SIGNALING PROTEIN-IMMUNE SYSTEM complex / MEMBRANE PROTEIN-SIGNALING PROTEIN complex | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationAdenylate cyclase inhibitory pathway / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / Extra-nuclear estrogen signaling / Muscarinic acetylcholine receptors / G protein-coupled acetylcholine receptor activity / G alpha (i) signalling events / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / regulation of locomotion / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger ...Adenylate cyclase inhibitory pathway / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / Extra-nuclear estrogen signaling / Muscarinic acetylcholine receptors / G protein-coupled acetylcholine receptor activity / G alpha (i) signalling events / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / regulation of locomotion / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / response to prostaglandin E / D2 dopamine receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / chemical synaptic transmission / Ras protein signal transduction / postsynaptic membrane / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / synapse / dendrite / centrosome / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / Golgi apparatus / signal transduction / extracellular exosome / nucleoplasm / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||
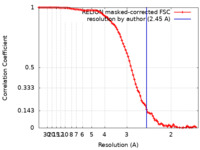
| Method | single particle reconstruction / cryo EM / Resolution: 2.45 Å | ||||||||||||||||||||||||
 Authors Authors | Vuckovic Z / Mobbs JI / Glukhova A / Sexton PM / Danev R / Thal DM | ||||||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Australia, Australia,  Japan, 7 items Japan, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Xanomeline displays concomitant orthosteric and allosteric binding modes at the M mAChR. Authors: Wessel A C Burger / Vi Pham / Ziva Vuckovic / Alexander S Powers / Jesse I Mobbs / Yianni Laloudakis / Alisa Glukhova / Denise Wootten / Andrew B Tobin / Patrick M Sexton / Steven M Paul / ...Authors: Wessel A C Burger / Vi Pham / Ziva Vuckovic / Alexander S Powers / Jesse I Mobbs / Yianni Laloudakis / Alisa Glukhova / Denise Wootten / Andrew B Tobin / Patrick M Sexton / Steven M Paul / Christian C Felder / Radostin Danev / Ron O Dror / Arthur Christopoulos / Celine Valant / David M Thal /     Abstract: The M muscarinic acetylcholine receptor (M mAChR) has emerged as a drug target of high therapeutic interest due to its expression in regions of the brain involved in the regulation of psychosis, ...The M muscarinic acetylcholine receptor (M mAChR) has emerged as a drug target of high therapeutic interest due to its expression in regions of the brain involved in the regulation of psychosis, cognition, and addiction. The mAChR agonist, xanomeline, has provided significant improvement in the Positive and Negative Symptom Scale (PANSS) scores in a Phase II clinical trial for the treatment of patients suffering from schizophrenia. Here we report the active state cryo-EM structure of xanomeline bound to the human M mAChR in complex with the heterotrimeric G transducer protein. Unexpectedly, two molecules of xanomeline were found to concomitantly bind to the monomeric M mAChR, with one molecule bound in the orthosteric (acetylcholine-binding) site and a second molecule in an extracellular vestibular allosteric site. Molecular dynamic simulations supports the structural findings, and pharmacological validation confirmed that xanomeline acts as a dual orthosteric and allosteric ligand at the human M mAChR. These findings provide a basis for further understanding xanomeline's complex pharmacology and highlight the myriad of ways through which clinically relevant ligands can bind to and regulate GPCRs. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29524.map.gz emd_29524.map.gz | 51.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29524-v30.xml emd-29524-v30.xml emd-29524.xml emd-29524.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29524_fsc.xml emd_29524_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_29524.png emd_29524.png | 127.1 KB | ||
| Masks |  emd_29524_msk_1.map emd_29524_msk_1.map | 67 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29524.cif.gz emd-29524.cif.gz | 7.4 KB | ||
| Others |  emd_29524_half_map_1.map.gz emd_29524_half_map_1.map.gz emd_29524_half_map_2.map.gz emd_29524_half_map_2.map.gz | 51.9 MB 52 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29524 http://ftp.pdbj.org/pub/emdb/structures/EMD-29524 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29524 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29524 | HTTPS FTP |
-Related structure data
| Related structure data |  8fx5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29524.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29524.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29524_msk_1.map emd_29524_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29524_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29524_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : M4 mAChR bound to agonist xanomeline in complex with dominant neg...
| Entire | Name: M4 mAChR bound to agonist xanomeline in complex with dominant negative Galpha-i1, Gbeta1, Ggamma2, and scFv16 |
|---|---|
| Components |
|
-Supramolecule #1: M4 mAChR bound to agonist xanomeline in complex with dominant neg...
| Supramolecule | Name: M4 mAChR bound to agonist xanomeline in complex with dominant negative Galpha-i1, Gbeta1, Ggamma2, and scFv16 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|
-Macromolecule #1: Muscarinic acetylcholine receptor M4
| Macromolecule | Name: Muscarinic acetylcholine receptor M4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.598277 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDAMA NFTPVNGSSG NQSVRLVTSS SHNRYETVEM VFIATVTGSL SLVTVVGNIL VMLSIKVNRQ LQTVNNYFLF SLACADLII GAFSMNLYTV YIIKGYWPLG AVVCDLWLAL DYVVSNASVM NLLIISFDRY FCVTKPLTYP ARRTTKMAGL M IAAAWVLS ...String: DYKDDDDAMA NFTPVNGSSG NQSVRLVTSS SHNRYETVEM VFIATVTGSL SLVTVVGNIL VMLSIKVNRQ LQTVNNYFLF SLACADLII GAFSMNLYTV YIIKGYWPLG AVVCDLWLAL DYVVSNASVM NLLIISFDRY FCVTKPLTYP ARRTTKMAGL M IAAAWVLS FVLWAPAILF WQFVVGKRTV PDNQCFIQFL SNPAVTFGTA IAAFYLPVVI MTVLYIHISL ASRSRVHKHR PE GPKEKKA KTKRQMAARE RKVTRTIFAI LLAFILTWTP YNVMVLVNTF CQSCIPDTVW SIGYWLCYVN STINPACYAL CNA TFKKTF RHLLLCQYRN IGTARHHHHH HHH UniProtKB: Muscarinic acetylcholine receptor M4, Muscarinic acetylcholine receptor M4 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.402867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHGSSG SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLI IWDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR F LDDNQIVT ...String: HHHHHHGSSG SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLI IWDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR F LDDNQIVT SSGDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NA ICFFPNG NAFATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGH DNRVSC LGVTDDGMAV ATGSWDSFLK IWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.729947 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: ASNNTASIAQ ARKLVEQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASENPF REKKFFCAIL UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Antibody fragment scFv16
| Macromolecule | Name: Antibody fragment scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.466486 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL K |
-Macromolecule #5: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.414047 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #6: xanomeline
| Macromolecule | Name: xanomeline / type: ligand / ID: 6 / Number of copies: 2 / Formula: XNO |
|---|---|
| Molecular weight | Theoretical: 281.417 Da |
| Chemical component information | 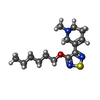 ChemComp-XNO: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 47 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 52.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)