[English] 日本語
 Yorodumi
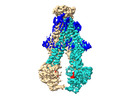
Yorodumi- EMDB-29362: Heterodimeric ABC transporter BmrCD in the inward-facing conforma... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to ATP: BmrCD_IF-ATP | |||||||||
 Map data Map data | Structure0524 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transporter / complex / lipids / MEMBRANE PROTEIN / TRANSLOCASE / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationATPase-coupled lipid transmembrane transporter activity / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / transmembrane transport / response to antibiotic / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | |||||||||
 Authors Authors | Tang Q / Mchaourab H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Asymmetric conformations and lipid interactions shape the ATP-coupled cycle of a heterodimeric ABC transporter. Authors: Qingyu Tang / Matt Sinclair / Hale S Hasdemir / Richard A Stein / Erkan Karakas / Emad Tajkhorshid / Hassane S Mchaourab /  Abstract: Here we used cryo-electron microscopy (cryo-EM), double electron-electron resonance spectroscopy (DEER), and molecular dynamics (MD) simulations, to capture and characterize ATP- and substrate-bound ...Here we used cryo-electron microscopy (cryo-EM), double electron-electron resonance spectroscopy (DEER), and molecular dynamics (MD) simulations, to capture and characterize ATP- and substrate-bound inward-facing (IF) and occluded (OC) conformational states of the heterodimeric ATP binding cassette (ABC) multidrug exporter BmrCD in lipid nanodiscs. Supported by DEER analysis, the structures reveal that ATP-powered isomerization entails changes in the relative symmetry of the BmrC and BmrD subunits that propagates from the transmembrane domain to the nucleotide binding domain. The structures uncover asymmetric substrate and Mg binding which we hypothesize are required for triggering ATP hydrolysis preferentially in one of the nucleotide-binding sites. MD simulations demonstrate that multiple lipid molecules differentially bind the IF versus the OC conformation thus establishing that lipid interactions modulate BmrCD energy landscape. Our findings are framed in a model that highlights the role of asymmetric conformations in the ATP-coupled transport with general implications to the mechanism of ABC transporters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29362.map.gz emd_29362.map.gz | 94.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29362-v30.xml emd-29362-v30.xml emd-29362.xml emd-29362.xml | 36 KB 36 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29362.png emd_29362.png | 71.8 KB | ||
| Filedesc metadata |  emd-29362.cif.gz emd-29362.cif.gz | 7.3 KB | ||
| Others |  emd_29362_additional_1.map.gz emd_29362_additional_1.map.gz emd_29362_additional_2.map.gz emd_29362_additional_2.map.gz emd_29362_additional_3.map.gz emd_29362_additional_3.map.gz emd_29362_additional_4.map.gz emd_29362_additional_4.map.gz emd_29362_additional_5.map.gz emd_29362_additional_5.map.gz emd_29362_additional_6.map.gz emd_29362_additional_6.map.gz emd_29362_half_map_1.map.gz emd_29362_half_map_1.map.gz emd_29362_half_map_2.map.gz emd_29362_half_map_2.map.gz | 94.5 MB 97.1 MB 97.2 MB 97.2 MB 97.2 MB 97.1 MB 95.2 MB 95.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29362 http://ftp.pdbj.org/pub/emdb/structures/EMD-29362 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29362 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29362 | HTTPS FTP |
-Related structure data
| Related structure data |  8fpfMC  8fhkC  8fmvC  8szcC  8t1pC  8t3kC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29362.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29362.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure0524 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
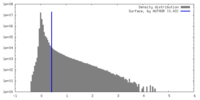
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Additional Map 1
| File | emd_29362_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Additional Map 2
| File | emd_29362_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Additional Map 3
| File | emd_29362_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map 3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Additional Map 4
| File | emd_29362_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map 4 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Additional Map 5
| File | emd_29362_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map 5 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Additional Map 6
| File | emd_29362_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map 6 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 1
| File | emd_29362_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_29362_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heterodimeric BmrCD with ATP in lipid bilayers
| Entire | Name: Heterodimeric BmrCD with ATP in lipid bilayers |
|---|---|
| Components |
|
-Supramolecule #1: Heterodimeric BmrCD with ATP in lipid bilayers
| Supramolecule | Name: Heterodimeric BmrCD with ATP in lipid bilayers / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Probable multidrug resistance ABC transporter ATP-binding/permeas...
| Macromolecule | Name: Probable multidrug resistance ABC transporter ATP-binding/permease protein YheH type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 77.369898 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKIGKTLWRY ALLYRKLLIT AVLLLTVAVG AELTGPFIGK KMIDDHILGI EKTWYEAAEK DKNAVQFHGV SYVREDRLQE PVSKAKEAH IYQVGMAFYF VDQAVSFDGN RTVSDGKLTI TNGDKSRAYA AEKLTKQELF QFYQPEIKGM VLLIALYGGL L VFSVFFQY ...String: MKIGKTLWRY ALLYRKLLIT AVLLLTVAVG AELTGPFIGK KMIDDHILGI EKTWYEAAEK DKNAVQFHGV SYVREDRLQE PVSKAKEAH IYQVGMAFYF VDQAVSFDGN RTVSDGKLTI TNGDKSRAYA AEKLTKQELF QFYQPEIKGM VLLIALYGGL L VFSVFFQY GQHYLLQMSA NRIIQKMRQD VFSHIQKMPI RYFDNLPAGK VVARITNDTE AIRDLYVTVL STFVTSGIYM FG IFTALFL LDVKLAFVAL AIVPIIWLWS VIYRRYASYY NQKIRSINSD INAKMNESIQ GMTIIQAFRH QKETMREFEE LNE SHFYFQ NRMLNLNSLM SHNLVNVIRN LAFVALIWHF GGASLNAAGI VSIGVLYAFV DYLNRLFQPI TGIVNQFSKL ELAR VSAGR VFELLEEKNT EEAGEPAKER ALGRVEFRDV SFAYQEGEEV LKHISFTAQK GETVALVGHT GSGKSSILNL LFRFY DAQK GDVLIDGKSI YNMSRQELRS HMGIVLQDPY LFSGTIGSNV SLDDERMTEE EIKNALRQVG AEPLLKKLPK GINEPV IEK GSTLSSGERQ LISFARALAF DPAILILDQA TAHIDTETEA VIQKALDVVK QGRTTFVIAH RLSTIRNADQ ILVLDKG EI VERGNHEELM ALEGQYYQMY ELQKGQKHSI ALEHHHHHH UniProtKB: Probable multidrug resistance ABC transporter ATP-binding/permease protein YheH |
-Macromolecule #2: Probable multidrug resistance ABC transporter ATP-binding/permeas...
| Macromolecule | Name: Probable multidrug resistance ABC transporter ATP-binding/permease protein YheI type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 67.602961 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MLEFSVLKKL GWFFKAYWLR YTIAIVLLLA VNVIEMFPPK LLGNAIDDMK AGAFTAEGLL FYIGIFFVL TAAVYIMSYF WMHQLFGGAN LMEKILRTKL MGHLLTMSPP FYEKNRTGDL MARGTNDLQA VSLTTGFGIL T LVDSTMFM ...String: MGSSHHHHHH SSGLVPRGSH MLEFSVLKKL GWFFKAYWLR YTIAIVLLLA VNVIEMFPPK LLGNAIDDMK AGAFTAEGLL FYIGIFFVL TAAVYIMSYF WMHQLFGGAN LMEKILRTKL MGHLLTMSPP FYEKNRTGDL MARGTNDLQA VSLTTGFGIL T LVDSTMFM MTIFLTMGFL ISWKLTFAAI IPLPVMAIAI SLYGSKIHER FTEAQNAFGA LNDRVLESVS GVRVIRAYVQ ET NDVRRFN EMTADVYQKN MKVAFIDSLF EPTVKLLVGA SYLIGLGYGA FLVFRNELTL GELVSFNVYL GMMIWPMFAI GEL INVMQR GNASLDRVNE TLSYETDVTD PKQPADLKEP GDIVFSHVSF TYPSSTSDNL QDISFTVRKG QTVGIAGKTG SGKT TIIKQ LLRQYPPGEG SITFSGVPIQ QIPLDRLRGW IGYVPQDHLL FSRTVKENIL YGKQDATDKE VQQAIAEAHF EKDLH MLPS GLETMVGEKG VALSGGQKQR ISIARALMAN PEILILDQSL SAVDAKTEAA IIKNIRENRK GKTTFILTHR LSAVEH ADL ILVMDGGVIA ERGTHQELLA NNGWYREQYE RQQLFTAEEG GAGA UniProtKB: Probable multidrug resistance ABC transporter ATP-binding/permease protein YheI |
-Macromolecule #3: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 3 / Number of copies: 25 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)