[English] 日本語
 Yorodumi
Yorodumi- EMDB-29295: Consensus structure of HIV-1 envelope trimer bound to CD4 recepto... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Consensus structure of HIV-1 envelope trimer bound to CD4 receptors on membranes | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Virus-Host Complex / Glycoprotein / Cell receptor / VIRAL PROTEIN | |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
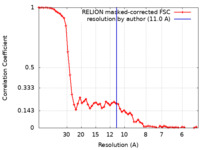
| Method | subtomogram averaging / cryo EM / Resolution: 11.0 Å | |||||||||
 Authors Authors | Li W / Qin Z / Nand E / Grunst MW / Grover JR / Uchil PD / Mothes W | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: HIV-1 Env trimers asymmetrically engage CD4 receptors in membranes. Authors: Wenwei Li / Zhuan Qin / Elizabeth Nand / Michael W Grunst / Jonathan R Grover / Julian W Bess / Jeffrey D Lifson / Michael B Zwick / Hemant D Tagare / Pradeep D Uchil / Walther Mothes /  Abstract: Human immunodeficiency virus 1 (HIV-1) infection is initiated by binding of the viral envelope glycoprotein (Env) to the cell-surface receptor CD4. Although high-resolution structures of Env in a ...Human immunodeficiency virus 1 (HIV-1) infection is initiated by binding of the viral envelope glycoprotein (Env) to the cell-surface receptor CD4. Although high-resolution structures of Env in a complex with the soluble domains of CD4 have been determined, the binding process is less understood in native membranes. Here we used cryo-electron tomography to monitor Env-CD4 interactions at the membrane-membrane interfaces formed between HIV-1 and CD4-presenting virus-like particles. Env-CD4 complexes organized into clusters and rings, bringing the opposing membranes closer together. Env-CD4 clustering was dependent on capsid maturation. Subtomogram averaging and classification revealed that Env bound to one, two and finally three CD4 molecules, after which Env adopted an open state. Our data indicate that asymmetric HIV-1 Env trimers bound to one and two CD4 molecules are detectable intermediates during virus binding to host cell membranes, which probably has consequences for antibody-mediated immune responses and vaccine immunogen design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29295.map.gz emd_29295.map.gz | 14.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29295-v30.xml emd-29295-v30.xml emd-29295.xml emd-29295.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29295_fsc.xml emd_29295_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_29295.png emd_29295.png | 54.6 KB | ||
| Filedesc metadata |  emd-29295.cif.gz emd-29295.cif.gz | 4.1 KB | ||
| Others |  emd_29295_half_map_1.map.gz emd_29295_half_map_1.map.gz emd_29295_half_map_2.map.gz emd_29295_half_map_2.map.gz | 14.4 MB 14.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29295 http://ftp.pdbj.org/pub/emdb/structures/EMD-29295 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29295 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29295 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29295.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29295.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.7 Å | ||||||||||||||||||||||||||||||||||||
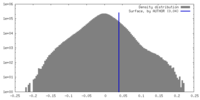
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29295_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29295_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Consensus structure of HIV-1 envelope trimer bound to CD4 recepto...
| Entire | Name: Consensus structure of HIV-1 envelope trimer bound to CD4 receptors on membranes |
|---|---|
| Components |
|
-Supramolecule #1: Consensus structure of HIV-1 envelope trimer bound to CD4 recepto...
| Supramolecule | Name: Consensus structure of HIV-1 envelope trimer bound to CD4 receptors on membranes type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.5 µm / Nominal defocus min: 0.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)