[English] 日本語
 Yorodumi
Yorodumi- EMDB-29279: Cryo-EM structure of a group II intron immediately before branching -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a group II intron immediately before branching | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | group II intron / splicing / branching / maturase / SPLICING-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA-directed DNA polymerase activity / endonuclease activity / nucleic acid binding / zinc ion binding Similarity search - Function | |||||||||
| Biological species |   Thermosynechococcus vestitus (bacteria) Thermosynechococcus vestitus (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Haack DB / Rudolfs BG / Zhang C / Lyumkis D / Toor N | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis of branching during RNA splicing. Authors: Daniel B Haack / Boris Rudolfs / Cheng Zhang / Dmitry Lyumkis / Navtej Toor /  Abstract: Branching is a critical step in RNA splicing that is essential for 5' splice site selection. Recent spliceosome structures have led to competing models for the recognition of the invariant adenosine ...Branching is a critical step in RNA splicing that is essential for 5' splice site selection. Recent spliceosome structures have led to competing models for the recognition of the invariant adenosine at the branch point. However, there are no structures of any splicing complex with the adenosine nucleophile docked in the active site and positioned to attack the 5' splice site. Thus we lack a mechanistic understanding of adenosine selection and splice site recognition during RNA splicing. Here we present a cryo-electron microscopy structure of a group II intron that reveals that active site dynamics are coupled to the formation of a base triple within the branch-site helix that positions the 2'-OH of the adenosine for nucleophilic attack on the 5' scissile phosphate. This structure, complemented with biochemistry and comparative analyses to splicing complexes, supports a base triple model of adenosine recognition for branching within group II introns and the evolutionarily related spliceosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29279.map.gz emd_29279.map.gz | 136.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29279-v30.xml emd-29279-v30.xml emd-29279.xml emd-29279.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29279.png emd_29279.png | 58.5 KB | ||
| Filedesc metadata |  emd-29279.cif.gz emd-29279.cif.gz | 6.1 KB | ||
| Others |  emd_29279_half_map_1.map.gz emd_29279_half_map_1.map.gz emd_29279_half_map_2.map.gz emd_29279_half_map_2.map.gz | 134.3 MB 134.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29279 http://ftp.pdbj.org/pub/emdb/structures/EMD-29279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29279 | HTTPS FTP |
-Related structure data
| Related structure data |  8fliMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29279.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29279.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
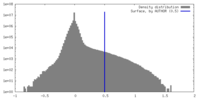
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29279_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
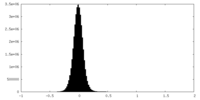
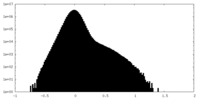
| Density Histograms |
-Half map: #1
| File | emd_29279_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
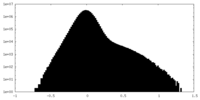
| Density Histograms |
- Sample components
Sample components
-Entire : Group IIB intron in complex with its maturase protein
| Entire | Name: Group IIB intron in complex with its maturase protein |
|---|---|
| Components |
|
-Supramolecule #1: Group IIB intron in complex with its maturase protein
| Supramolecule | Name: Group IIB intron in complex with its maturase protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus vestitus (bacteria) Thermosynechococcus vestitus (bacteria) |
-Macromolecule #1: Group II Intron
| Macromolecule | Name: Group II Intron / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus vestitus (bacteria) Thermosynechococcus vestitus (bacteria) |
| Molecular weight | Theoretical: 289.257938 KDa |
| Sequence | String: CCCAGGGUUG GCCGAGCGUU GCGACGCGAA AGCUAGCCAG AUGAUUGUCC CACUAGCCCA ACAAGCUAGA ACGGGACCGG UUGUUCCCC CAACCGUAGC CUAGGGAGGC AUGCGUGACU GGUAACGGUC AGGUGUGAAG CCCUCCCGAC AAUGUAGCCC G AACCGCAA ...String: CCCAGGGUUG GCCGAGCGUU GCGACGCGAA AGCUAGCCAG AUGAUUGUCC CACUAGCCCA ACAAGCUAGA ACGGGACCGG UUGUUCCCC CAACCGUAGC CUAGGGAGGC AUGCGUGACU GGUAACGGUC AGGUGUGAAG CCCUCCCGAC AAUGUAGCCC G AACCGCAA GGUUGAAGCU GAAUCCGUGA GGAGGAAGCA ACUUCACCAG UGUCAGGUGA UAGGGAACUA GGCUUGAGGG UA UGGUGAG CACAUGCGAA GUGAUGUCAG AAGCCUCGUC ACAGACCAAC AGGCCAAAGA CACUGAUAGG CCUGAGCCAA AAC GGCAAA UGGAUAGGCU ACAUCGCUCG CUCGUCGGUG UACGGGGACG UCAAUCCAUC GGGGCACAGU CACCACCUAA CCCC UCGUG UCAUCUGGUU GGAACGCGGU AAGCCCGUAU CCUCGCCUUG AACACUCAAG GCAGGCAAAC CUUCGGGAAU GCUGA UGGG GGUGCGGGUA UGGGAUGCAG GAGAAAGCGA AUGCCGGUCU GUAAUGGACC GGAUAGGGGU UGAGGAGACA AUCCAA CAU CACCCCGCCC GAAAGGGAGC AGACUUCCUG CUGGUCUCUC UUUGCGAGAU AGCCUGUAGA ACCUCUUGAA UGGAGAC AA GGCAAAUGGC AGUGGAACAA ACCACUGGUG CGGUCACCAA CCAAACGGAA ACAAGCUGGC ACAGCAUAGA CUGGGCCA A AGCCAACCGU GAGGUAAAGA GGCUGCAAGU GCGUAUCGCA AAGGCGUUCG CGCCGGUUCC UCUUGAAAGA GGGGCUUUG AGAGGCCUGA GCCGGAUGUG GGGAAACUCA CAAGUCCGGU UCUUAGGGGG CGGGGAUGGC AUUCGUGCCU CCCUGCUACC CGGCGAUGA GGCA |
-Macromolecule #2: Maturase reverse transcriptase
| Macromolecule | Name: Maturase reverse transcriptase / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermosynechococcus vestitus (bacteria) Thermosynechococcus vestitus (bacteria) |
| Molecular weight | Theoretical: 65.065121 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: METRQMAVEQ TTGAVTNQTE TSWHSIDWAK ANREVKRLQV RIAKAVKEGR WGKVKALQWL LTHSFYGKAL AVKRVTDNSG SKTPGVDGI TWSTQEQKAQ AIKSLRRRGY KPQPLRRVYI PKASGKQRPL GIPTTKDRAM QALYALALEP VAETTADRNS Y GFRQGRCT ...String: METRQMAVEQ TTGAVTNQTE TSWHSIDWAK ANREVKRLQV RIAKAVKEGR WGKVKALQWL LTHSFYGKAL AVKRVTDNSG SKTPGVDGI TWSTQEQKAQ AIKSLRRRGY KPQPLRRVYI PKASGKQRPL GIPTTKDRAM QALYALALEP VAETTADRNS Y GFRQGRCT ADAAGQCFTV LGRSDCAKYI LDADITGCFD NISHEWLLDN IPLDKEVLRK WLKSGFVWKQ QLFPTHAGTP QG GVISPML ANMTLDGMEE LLKKHLRKQK VNLIRYADDF VVTGESKETL EKVTTVIQEF LKERGLTLSE EKTKVVHIEE GFD FLGWNI RKYGEKLLIK PAKKNIKAFH KKIRDALKEL RTATQEAVID TLNPIIKGWA NYHRNQVSKR IFNRADDNIW HKLW RWAKR RHPNKPARWT KNKYFIKIGN RHWVFGTWKK DKEGRLRSRY LIKAGDTRIQ RHVKIKADAN PFLPEWAEYF EERKK LKEA PAQYRRIRRE LWKKQGGICP VCGGEIEQDM LTEIHHILPK HKGGSDDLDN LVLIHANCHK QVHSRDGQHS RFLLKE GL UniProtKB: Maturase reverse transcriptase |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 31.6 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 38881 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)