[English] 日本語
 Yorodumi
Yorodumi- EMDB-29221: CryoEM structure of HLA-A2 MAGEA4 (286-294) in complex with H2aM3... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of HLA-A2 MAGEA4 (286-294) in complex with H2aM31345N Fab and 2M2 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HLA / MHC / IMMUNE SYSTEM / antibody | |||||||||
| Function / homology |  Function and homology information Function and homology informationantigen processing and presentation of peptide antigen via MHC class I / early endosome lumen / Nef mediated downregulation of MHC class I complex cell surface expression / DAP12 interactions / Endosomal/Vacuolar pathway / Antigen Presentation: Folding, assembly and peptide loading of class I MHC / negative regulation of iron ion transport / lumenal side of endoplasmic reticulum membrane / T cell mediated cytotoxicity / cellular response to iron(III) ion ...antigen processing and presentation of peptide antigen via MHC class I / early endosome lumen / Nef mediated downregulation of MHC class I complex cell surface expression / DAP12 interactions / Endosomal/Vacuolar pathway / Antigen Presentation: Folding, assembly and peptide loading of class I MHC / negative regulation of iron ion transport / lumenal side of endoplasmic reticulum membrane / T cell mediated cytotoxicity / cellular response to iron(III) ion / negative regulation of forebrain neuron differentiation / antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent / ER to Golgi transport vesicle membrane / peptide antigen assembly with MHC class I protein complex / transferrin transport / regulation of iron ion transport / regulation of erythrocyte differentiation / negative regulation of receptor-mediated endocytosis / HFE-transferrin receptor complex / response to molecule of bacterial origin / MHC class I peptide loading complex / cellular response to iron ion / positive regulation of T cell cytokine production / antigen processing and presentation of endogenous peptide antigen via MHC class I / MHC class I protein complex / peptide antigen assembly with MHC class II protein complex / negative regulation of neurogenesis / positive regulation of receptor-mediated endocytosis / cellular response to nicotine / MHC class II protein complex / positive regulation of T cell mediated cytotoxicity / multicellular organismal-level iron ion homeostasis / specific granule lumen / peptide antigen binding / antigen processing and presentation of exogenous peptide antigen via MHC class II / phagocytic vesicle membrane / positive regulation of immune response / recycling endosome membrane / histone deacetylase binding / positive regulation of T cell activation / Interferon gamma signaling / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / negative regulation of epithelial cell proliferation / Modulation by Mtb of host immune system / sensory perception of smell / positive regulation of cellular senescence / tertiary granule lumen / DAP12 signaling / MHC class II protein complex binding / T cell differentiation in thymus / late endosome membrane / negative regulation of neuron projection development / ER-Phagosome pathway / protein refolding / early endosome membrane / amyloid fibril formation / protein homotetramerization / intracellular iron ion homeostasis / learning or memory / immune response / endoplasmic reticulum lumen / Amyloid fiber formation / Golgi membrane / external side of plasma membrane / lysosomal membrane / focal adhesion / Neutrophil degranulation / SARS-CoV-2 activates/modulates innate and adaptive immune responses / structural molecule activity / cell surface / negative regulation of transcription by RNA polymerase II / endoplasmic reticulum / Golgi apparatus / protein homodimerization activity / extracellular space / extracellular exosome / extracellular region / metal ion binding / identical protein binding / membrane / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Saotome K / Franklin MC | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2025 Journal: Sci Transl Med / Year: 2025Title: CAR T cells based on fully human T cell receptor-mimetic antibodies exhibit potent antitumor activity in vivo. Authors: Robert Salzler / David J DiLillo / Kei Saotome / Kevin Bray / Katja Mohrs / Haun Hwang / Kamil J Cygan / Darshit Shah / Anna Rye-Weller / Kunal Kundu / Ashok Badithe / Xiaoqin Zhang / Elena ...Authors: Robert Salzler / David J DiLillo / Kei Saotome / Kevin Bray / Katja Mohrs / Haun Hwang / Kamil J Cygan / Darshit Shah / Anna Rye-Weller / Kunal Kundu / Ashok Badithe / Xiaoqin Zhang / Elena Garnova / Marcela Torres / Ankur Dhanik / Robert Babb / Frank J Delfino / Courtney Thwaites / Drew Dudgeon / Michael J Moore / Thomas Craig Meagher / Corinne E Decker / Tomasz Owczarek / John A Gleason / Xiaoran Yang / David Suh / Wen-Yi Lee / Richard Welsh / Douglas MacDonald / Johanna Hansen / Chunguang Guo / Jessica R Kirshner / Gavin Thurston / Tammy Huang / Matthew C Franklin / George D Yancopoulos / John C Lin / Lynn E Macdonald / Andrew J Murphy / Gang Chen / Olav Olsen / William C Olson /  Abstract: Monoclonal antibody therapies have transformed the lives of patients across a diverse range of diseases. However, antibodies can usually only access extracellular proteins, including the ...Monoclonal antibody therapies have transformed the lives of patients across a diverse range of diseases. However, antibodies can usually only access extracellular proteins, including the extracellular portions of membrane proteins that are expressed on the cell surface. In contrast, T cell receptors (TCRs) survey the entire cellular proteome when processed and presented as peptides in association with human leukocyte antigen (pHLA complexes). Antibodies that mimic TCRs by recognizing pHLA complexes have the potential to extend the reach of antibodies to this larger pool of targets and provide increased binding affinity and specificity. A major challenge in developing TCR mimetic (TCRm) antibodies is the limited sequence differences between the target pHLA complex relative to the large global repertoire of pHLA complexes. Here, we provide a comprehensive strategy for generating fully human TCRm antibodies across multiple HLA alleles, beginning with pHLA target discovery and validation and culminating in the engineering of TCRm-based chimeric antigen receptor T cells with potent antitumor activity. By incorporating mass spectrometry, bioinformatic predictions, HLA-humanized mice, antibody screening, and cryo-electron microscopy, we have established a pipeline to identify additional pHLA complex-specific antibodies with therapeutic potential. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29221.map.gz emd_29221.map.gz | 147.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29221-v30.xml emd-29221-v30.xml emd-29221.xml emd-29221.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29221.png emd_29221.png | 83.7 KB | ||
| Filedesc metadata |  emd-29221.cif.gz emd-29221.cif.gz | 7.1 KB | ||
| Others |  emd_29221_half_map_1.map.gz emd_29221_half_map_1.map.gz emd_29221_half_map_2.map.gz emd_29221_half_map_2.map.gz | 141 MB 140.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29221 http://ftp.pdbj.org/pub/emdb/structures/EMD-29221 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29221 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29221 | HTTPS FTP |
-Related structure data
| Related structure data |  8fjbMC  8fjaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29221.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29221.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
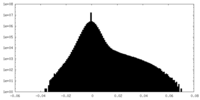
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
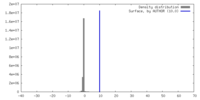
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29221_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
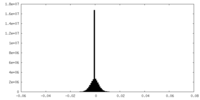
| Density Histograms |
-Half map: #2
| File | emd_29221_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
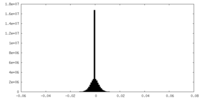
| Density Histograms |
- Sample components
Sample components
-Entire : HLA-A2 MAGEA4 (286-294) complex with H2aM31345N Fab and 2M2 Fab
| Entire | Name: HLA-A2 MAGEA4 (286-294) complex with H2aM31345N Fab and 2M2 Fab |
|---|---|
| Components |
|
-Supramolecule #1: HLA-A2 MAGEA4 (286-294) complex with H2aM31345N Fab and 2M2 Fab
| Supramolecule | Name: HLA-A2 MAGEA4 (286-294) complex with H2aM31345N Fab and 2M2 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: MHC class I antigen, Beta-2-microglobulin
| Supramolecule | Name: MHC class I antigen, Beta-2-microglobulin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Melanoma-associated antigen 4 peptide
| Supramolecule | Name: Melanoma-associated antigen 4 peptide / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|
-Supramolecule #4: H2aM31345N Fab heavy chain, H2aM31345N Fab light chain
| Supramolecule | Name: H2aM31345N Fab heavy chain, H2aM31345N Fab light chain type: complex / ID: 4 / Parent: 1 / Macromolecule list: #4-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: MHC class I antigen
| Macromolecule | Name: MHC class I antigen / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.082512 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSHSMRYFF TSVSRPGRGE PRFIAVGYVD DTQFVRFDSD AASQRMEPRA PWIEQEGPEY WDGETRKVKA HSQTHRVDLG TLRGYYNQS EAGSHTVQRM YGCDVGSDWR FLRGYHQYAY DGKDYIALKE DLRSWTAADM AAQTTKHKWE AAHVAEQLRA Y LEGTCVEW ...String: MGSHSMRYFF TSVSRPGRGE PRFIAVGYVD DTQFVRFDSD AASQRMEPRA PWIEQEGPEY WDGETRKVKA HSQTHRVDLG TLRGYYNQS EAGSHTVQRM YGCDVGSDWR FLRGYHQYAY DGKDYIALKE DLRSWTAADM AAQTTKHKWE AAHVAEQLRA Y LEGTCVEW LRRYLENGKE TLQRTDAPKT HMTHHAVSDH EATLRCWALS FYPAEITLTW QRDGEDQTQD TELVETRPAG DG TFQKWAA VVVPSGQEQR YTCHVQHEGL PKPLTLRWEP UniProtKB: MHC class I antigen |
-Macromolecule #2: Beta-2-microglobulin
| Macromolecule | Name: Beta-2-microglobulin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.879356 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIQRTPKIQV YSRHPAENGK SNFLNCYVSG FHPSDIEVDL LKNGERIEKV EHSDLSFSKD WSFYLLYYTE FTPTEKDEYA CRVNHVTLS QPKIVKWDRD M UniProtKB: Beta-2-microglobulin |
-Macromolecule #3: Melanoma-associated antigen 4 peptide
| Macromolecule | Name: Melanoma-associated antigen 4 peptide / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.08133 KDa |
| Sequence | String: KVLEHVVRV UniProtKB: Melanoma-associated antigen 8 |
-Macromolecule #4: H2aM31345N Fab heavy chain
| Macromolecule | Name: H2aM31345N Fab heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.831629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVESGGG LVKPGGSLRL SCAASGFTFS EYYMTWIRQA PGQGLEWVSY ISSSGFNIYY ADSVKGRFTI SRDNAKNSLF LQMNSLRVE DTAVYYCARE GVTDGMDVWG QGTTVTVSSA STKGPSVFPL APCSRSTSES TAALGCLVKD YFPEPVTVSW N SGALTSGV ...String: QVQLVESGGG LVKPGGSLRL SCAASGFTFS EYYMTWIRQA PGQGLEWVSY ISSSGFNIYY ADSVKGRFTI SRDNAKNSLF LQMNSLRVE DTAVYYCARE GVTDGMDVWG QGTTVTVSSA STKGPSVFPL APCSRSTSES TAALGCLVKD YFPEPVTVSW N SGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTKT YTCNVDHKPS NTKVDKRVES KYGPP |
-Macromolecule #5: H2aM31345N Fab light chain
| Macromolecule | Name: H2aM31345N Fab light chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.438967 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQSIS SYLNWYQQKP GKAPKLLIYA ASSLQSGVPS RFSGSGSGTD FTLTISSLQP EDFATYYCQ QSYSTPPITF GQGTRLEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: DIQMTQSPSS LSASVGDRVT ITCRASQSIS SYLNWYQQKP GKAPKLLIYA ASSLQSGVPS RFSGSGSGTD FTLTISSLQP EDFATYYCQ QSYSTPPITF GQGTRLEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)