[English] 日本語
 Yorodumi
Yorodumi- EMDB-2900: Structures of the CRISPR-Cmr complex reveal mode of RNA target po... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2900 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
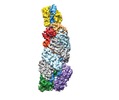
| Title | Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning | |||||||||
 Map data Map data | Reconstruction of target-bound CRISPR-Cas complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of defense response to virus / defense response to virus / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Taylor DW / Zhu Y / Staals RHJ / Kornfeld JE / Shinkai A / van der Oost J / Nogales E / Doudna JA | |||||||||
 Citation Citation |  Journal: Science / Year: 2015 Journal: Science / Year: 2015Title: Structural biology. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Authors: David W Taylor / Yifan Zhu / Raymond H J Staals / Jack E Kornfeld / Akeo Shinkai / John van der Oost / Eva Nogales / Jennifer A Doudna /    Abstract: Adaptive immunity in bacteria involves RNA-guided surveillance complexes that use CRISPR (clustered regularly interspaced short palindromic repeats)-associated (Cas) proteins together with CRISPR ...Adaptive immunity in bacteria involves RNA-guided surveillance complexes that use CRISPR (clustered regularly interspaced short palindromic repeats)-associated (Cas) proteins together with CRISPR RNAs (crRNAs) to target invasive nucleic acids for degradation. Whereas type I and type II CRISPR-Cas surveillance complexes target double-stranded DNA, type III complexes target single-stranded RNA. Near-atomic resolution cryo-electron microscopy reconstructions of native type III Cmr (CRISPR RAMP module) complexes in the absence and presence of target RNA reveal a helical protein arrangement that positions the crRNA for substrate binding. Thumblike β hairpins intercalate between segments of duplexed crRNA:target RNA to facilitate cleavage of the target at 6-nucleotide intervals. The Cmr complex is architecturally similar to the type I CRISPR-Cascade complex, suggesting divergent evolution of these immune systems from a common ancestor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2900.map.gz emd_2900.map.gz | 116.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2900-v30.xml emd-2900-v30.xml emd-2900.xml emd-2900.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2900.png emd_2900.png | 479.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2900 http://ftp.pdbj.org/pub/emdb/structures/EMD-2900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2900 | HTTPS FTP |
-Validation report
| Summary document |  emd_2900_validation.pdf.gz emd_2900_validation.pdf.gz | 288.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2900_full_validation.pdf.gz emd_2900_full_validation.pdf.gz | 287.6 KB | Display | |
| Data in XML |  emd_2900_validation.xml.gz emd_2900_validation.xml.gz | 5.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2900 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2900 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2900 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2900 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2900.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2900.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of target-bound CRISPR-Cas complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : target-bound CRISPR-Cmr complex
| Entire | Name: target-bound CRISPR-Cmr complex |
|---|---|
| Components |
|
-Supramolecule #1000: target-bound CRISPR-Cmr complex
| Supramolecule | Name: target-bound CRISPR-Cmr complex / type: sample / ID: 1000 / Number unique components: 13 |
|---|---|
| Molecular weight | Experimental: 381 KDa / Method: MS/MS |
-Macromolecule #1: CRISPR-associated protein TM1791
| Macromolecule | Name: CRISPR-associated protein TM1791 / type: protein_or_peptide / ID: 1 / Name.synonym: Cmr6, TTHB165 / Number of copies: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Experimental: 38 KDa / Theoretical: 38 KDa |
| Sequence | UniProtKB: CRISPR type III-associated protein domain-containing protein InterPro: CRISPR-associated protein, TM1791 |
-Macromolecule #4: CRISPR system Cmr subunit Cmr5
| Macromolecule | Name: CRISPR system Cmr subunit Cmr5 / type: protein_or_peptide / ID: 4 Name.synonym: Cmr5, CRISPR type III-B/RAMP module-associated protein Cmr5 Number of copies: 3 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Experimental: 12 KDa / Theoretical: 12 KDa |
| Sequence | UniProtKB: CRISPR system Cmr subunit Cmr5 / GO: regulation of defense response to virus / InterPro: CRISPR-associated protein, Cmr5 |
-Macromolecule #5: CRISPR-associated protein Cmr3
| Macromolecule | Name: CRISPR-associated protein Cmr3 / type: protein_or_peptide / ID: 5 / Name.synonym: Cmr3, TTHB160 / Number of copies: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Experimental: 39 KDa / Theoretical: 39 KDa |
| Sequence | UniProtKB: CRISPR-associated protein Cmr3 / InterPro: CRISPR-associated protein, Cmr3 |
-Macromolecule #6: CRISPR-associated protein Crm2
| Macromolecule | Name: CRISPR-associated protein Crm2 / type: protein_or_peptide / ID: 6 / Name.synonym: Cmr2, TTHB160 / Number of copies: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Experimental: 65 KDa / Theoretical: 65 KDa |
| Sequence | UniProtKB: GGDEF domain-containing protein / InterPro: CRISPR-associated protein Cmr2 |
-Macromolecule #7: CRISPR-associated RAMP Cmr4
| Macromolecule | Name: CRISPR-associated RAMP Cmr4 / type: protein_or_peptide / ID: 7 / Name.synonym: Cmr4, TTHB163 / Number of copies: 4 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Experimental: 37 KDa / Theoretical: 37 KDa |
| Sequence | UniProtKB: CRISPR type III-associated protein domain-containing protein InterPro: CRISPR-associated RAMP Cmr4 |
-Macromolecule #8: CRISPR-associated protein TM1795
| Macromolecule | Name: CRISPR-associated protein TM1795 / type: protein_or_peptide / ID: 8 / Name.synonym: Cmr1, TTHB162 / Number of copies: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Experimental: 44 KDa |
| Sequence | UniProtKB: CRISPR type III-associated protein domain-containing protein InterPro: CRISPR-associated protein TM1795 |
-Macromolecule #2: target RNA
| Macromolecule | Name: target RNA / type: rna / ID: 2 / Details: 50 nucleotide target RNA / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Theoretical: 16.2 KDa |
-Macromolecule #3: CRISPR RNA
| Macromolecule | Name: CRISPR RNA / type: rna / ID: 3 / Name.synonym: crRNA / Details: 46 nucleotide endogenous crRNA bound to complex / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris-HCl, pH 8.0, 150 mM NaCl |
| Grid | Details: 4/2 C-flat grids with a thin-layer of carbon over the holes |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 100 K / Instrument: FEI VITROBOT MARK IV Method: Grids were rapidly plunged into liquid ethane using an FEI Vitrobot MarkIV maintained at 4 degrees C after being blotted for 4-4.5 seconds with a blotting force of 15-20. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective astigmatism was corrected at 210,000 times magnification |
| Details | Data acquired using Leginon. We collected a 6 s exposure fractionated into 20, 300 ms frames with a dose of 8 electrons per square Angstrom per second. |
| Date | Jul 31, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Number real images: 4000 / Average electron dose: 48 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -4.5 µm / Nominal defocus min: -2.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: CTFFind3 |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 4.4 Å / Resolution method: OTHER / Software - Name: Relion / Number images used: 175000 |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















