+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human TRPV4 - RhoA, apo condition | |||||||||
 Map data Map data | Main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRPV4 / RhoA / MEMBRANE PROTEIN / MEMBRANE PROTEIN-Hydrolase complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationstretch-activated, monoatomic cation-selective, calcium channel activity / blood vessel endothelial cell delamination / regulation of response to osmotic stress / osmosensor activity / vasopressin secretion / calcium ion import into cytosol / positive regulation of microtubule depolymerization / positive regulation of striated muscle contraction / positive regulation of macrophage inflammatory protein 1 alpha production / hyperosmotic salinity response ...stretch-activated, monoatomic cation-selective, calcium channel activity / blood vessel endothelial cell delamination / regulation of response to osmotic stress / osmosensor activity / vasopressin secretion / calcium ion import into cytosol / positive regulation of microtubule depolymerization / positive regulation of striated muscle contraction / positive regulation of macrophage inflammatory protein 1 alpha production / hyperosmotic salinity response / positive regulation of chemokine (C-X-C motif) ligand 1 production / positive regulation of chemokine (C-C motif) ligand 5 production / cartilage development involved in endochondral bone morphogenesis / alpha-beta T cell lineage commitment / aortic valve formation / mitotic cleavage furrow formation / positive regulation of lipase activity / bone trabecula morphogenesis / endothelial tube lumen extension / skeletal muscle satellite cell migration / positive regulation of vascular associated smooth muscle contraction / angiotensin-mediated vasoconstriction involved in regulation of systemic arterial blood pressure / SLIT2:ROBO1 increases RHOA activity / RHO GTPases Activate Rhotekin and Rhophilins / Roundabout signaling pathway / negative regulation of intracellular steroid hormone receptor signaling pathway / Axonal growth inhibition (RHOA activation) / Axonal growth stimulation / cellular hypotonic salinity response / cleavage furrow formation / cellular hypotonic response / regulation of neural precursor cell proliferation / regulation of osteoblast proliferation / cortical microtubule organization / regulation of modification of postsynaptic actin cytoskeleton / forebrain radial glial cell differentiation / multicellular organismal-level water homeostasis / apical junction assembly / regulation of modification of postsynaptic structure / negative regulation of cell migration involved in sprouting angiogenesis / cell junction assembly / negative regulation of brown fat cell differentiation / positive regulation of vascular permeability / beta selection / establishment of epithelial cell apical/basal polarity / cellular response to chemokine / negative regulation of motor neuron apoptotic process / regulation of systemic arterial blood pressure by endothelin / negative regulation of oxidative phosphorylation / calcium ion import / osmosensory signaling pathway / RHO GTPases Activate ROCKs / RHO GTPases activate CIT / negative regulation of cell size / positive regulation of monocyte chemotactic protein-1 production / cell-cell junction assembly / Sema4D induced cell migration and growth-cone collapse / PCP/CE pathway / RHO GTPases activate KTN1 / cellular response to osmotic stress / positive regulation of podosome assembly / cell volume homeostasis / positive regulation of alpha-beta T cell differentiation / apolipoprotein A-I-mediated signaling pathway / Sema4D mediated inhibition of cell attachment and migration / wound healing, spreading of cells / positive regulation of leukocyte adhesion to vascular endothelial cell / PI3K/AKT activation / Wnt signaling pathway, planar cell polarity pathway / motor neuron apoptotic process / odontogenesis / ossification involved in bone maturation / regulation of focal adhesion assembly / regulation of aerobic respiration / negative chemotaxis / EPHA-mediated growth cone collapse / TRP channels / apical junction complex / androgen receptor signaling pathway / cortical actin cytoskeleton / diet induced thermogenesis / stress fiber assembly / myosin binding / positive regulation of cytokinesis / RHOC GTPase cycle / regulation of neuron projection development / positive regulation of macrophage chemotaxis / cellular response to cytokine stimulus / cerebral cortex cell migration / microtubule polymerization / ERBB2 Regulates Cell Motility / cleavage furrow / calcium ion import across plasma membrane / semaphorin-plexin signaling pathway / positive regulation of protein serine/threonine kinase activity / ficolin-1-rich granule membrane / negative regulation of cell-substrate adhesion / RHOA GTPase cycle / mitotic spindle assembly / response to mechanical stimulus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.75 Å | |||||||||
 Authors Authors | Kwon DH / Lee S-Y / Zhang F | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease. Authors: Do Hoon Kwon / Feng Zhang / Brett A McCray / Shasha Feng / Meha Kumar / Jeremy M Sullivan / Wonpil Im / Charlotte J Sumner / Seok-Yong Lee /  Abstract: Crosstalk between ion channels and small GTPases is critical during homeostasis and disease, but little is known about the structural underpinnings of these interactions. TRPV4 is a polymodal, ...Crosstalk between ion channels and small GTPases is critical during homeostasis and disease, but little is known about the structural underpinnings of these interactions. TRPV4 is a polymodal, calcium-permeable cation channel that has emerged as a potential therapeutic target in multiple conditions. Gain-of-function mutations also cause hereditary neuromuscular disease. Here, we present cryo-EM structures of human TRPV4 in complex with RhoA in the ligand-free, antagonist-bound closed, and agonist-bound open states. These structures reveal the mechanism of ligand-dependent TRPV4 gating. Channel activation is associated with rigid-body rotation of the intracellular ankyrin repeat domain, but state-dependent interaction with membrane-anchored RhoA constrains this movement. Notably, many residues at the TRPV4-RhoA interface are mutated in disease and perturbing this interface by introducing mutations into either TRPV4 or RhoA increases TRPV4 channel activity. Together, these results suggest that RhoA serves as an auxiliary subunit for TRPV4, regulating TRPV4-mediated calcium homeostasis and disruption of TRPV4-RhoA interactions can lead to TRPV4-related neuromuscular disease. These insights will help facilitate TRPV4 therapeutics development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28977.map.gz emd_28977.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28977-v30.xml emd-28977-v30.xml emd-28977.xml emd-28977.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28977.png emd_28977.png | 78.5 KB | ||
| Filedesc metadata |  emd-28977.cif.gz emd-28977.cif.gz | 6.7 KB | ||
| Others |  emd_28977_half_map_1.map.gz emd_28977_half_map_1.map.gz emd_28977_half_map_2.map.gz emd_28977_half_map_2.map.gz | 58.9 MB 58.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28977 http://ftp.pdbj.org/pub/emdb/structures/EMD-28977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28977 | HTTPS FTP |
-Related structure data
| Related structure data |  8fc9MC  8fc7C  8fc8C  8fcaC  8fcbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28977.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28977.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half B
| File | emd_28977_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
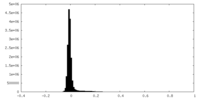
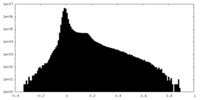
| Density Histograms |
-Half map: Half A
| File | emd_28977_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
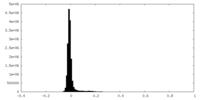
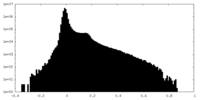
| Density Histograms |
- Sample components
Sample components
-Entire : The complex of human TRPV4 with RhoA
| Entire | Name: The complex of human TRPV4 with RhoA |
|---|---|
| Components |
|
-Supramolecule #1: The complex of human TRPV4 with RhoA
| Supramolecule | Name: The complex of human TRPV4 with RhoA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 450 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 4
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 4 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 102.057797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADSSEGPRA GPGEVAELPG DESGTPGGEA FPLSSLANLF EGEDGSLSPS PADASRPAGP GDGRPNLRMK FQGAFRKGVP NPIDLLEST LYESSVVPGP KKAPMDSLFD YGTYRHHSSD NKRWRKKIIE KQPQSPKAPA PQPPPILKVF NRPILFDIVS R GSTADLDG ...String: MADSSEGPRA GPGEVAELPG DESGTPGGEA FPLSSLANLF EGEDGSLSPS PADASRPAGP GDGRPNLRMK FQGAFRKGVP NPIDLLEST LYESSVVPGP KKAPMDSLFD YGTYRHHSSD NKRWRKKIIE KQPQSPKAPA PQPPPILKVF NRPILFDIVS R GSTADLDG LLPFLLTHKK RLTDEEFREP STGKTCLPKA LLNLSNGRND TIPVLLDIAE RTGNMREFIN SPFRDIYYRG QT ALHIAIE RRCKHYVELL VAQGADVHAQ ARGRFFQPKD EGGYFYFGEL PLSLAACTNQ PHIVNYLTEN PHKKADMRRQ DSR GNTVLH ALVAIADNTR ENTKFVTKMY DLLLLKCARL FPDSNLEAVL NNDGLSPLMM AAKTGKIGIF QHIIRREVTD EDTR HLSRK FKDWAYGPVY SSLYDLSSLD TCGEEASVLE ILVYNSKIEN RHEMLAVEPI NELLRDKWRK FGAVSFYINV VSYLC AMVI FTLTAYYQPL EGTPPYPYRT TVDYLRLAGE VITLFTGVLF FFTNIKDLFM KKCPGVNSLF IDGSFQLLYF IYSVLV IVS AALYLAGIEA YLAVMVFALV LGWMNALYFT RGLKLTGTYS IMIQKILFKD LFRFLLVYLL FMIGYASALV SLLNPCA NM KVCNEDQTNC TVPTYPSCRD SETFSTFLLD LFKLTIGMGD LEMLSSTKYP VVFIILLVTY IILTFVLLLN MLIALMGE T VGQVSKESKH IWKLQWATTI LDIERSFPVF LRKAFRSGEM VTVGKSSDGT PDRRWCFRVD EVNWSHWNQN LGIINEDPG KNETYQYYGF SHTVGRLRRD RWSSVVPRVV ELNKNSNPDE VVVPLDSMGN PRCDGHQQGY PRKWRTDDAP LENSLEVLFQ GPDYKDDDD KAHHHHHHHH HH UniProtKB: Transient receptor potential cation channel subfamily V member 4 |
-Macromolecule #2: Transforming protein RhoA
| Macromolecule | Name: Transforming protein RhoA / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: small monomeric GTPase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 21.799158 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAIRKKLVI VGDGACGKTC LLIVFSKDQF PEVYVPTVFE NYVADIEVDG KQVELALWDT AGQEDYDRLR PLSYPDTDVI LMCFSIDSP DSLENIPEKW TPEVKHFCPN VPIILVGNKK DLRNDEHTRR ELAKMKQEPV KPEEGRDMAN RIGAFGYMEC S AKTKDGVR ...String: MAAIRKKLVI VGDGACGKTC LLIVFSKDQF PEVYVPTVFE NYVADIEVDG KQVELALWDT AGQEDYDRLR PLSYPDTDVI LMCFSIDSP DSLENIPEKW TPEVKHFCPN VPIILVGNKK DLRNDEHTRR ELAKMKQEPV KPEEGRDMAN RIGAFGYMEC S AKTKDGVR EVFEMATRAA LQARRGKKKS GCLVL UniProtKB: Transforming protein RhoA |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 4 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)