[English] 日本語
 Yorodumi
Yorodumi- EMDB-28936: Cryo-EM structure of the Tropomodulin-capped pointed end of F-actin -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Tropomodulin-capped pointed end of F-actin | ||||||||||||
 Map data Map data | Final map of the Tropomodulin-bound actin filament pointed end. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Actin cytoskeleton / filament / Tmod / STRUCTURAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpointed-end actin filament capping / lens fiber cell development / myofibril assembly / Striated Muscle Contraction / cytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / myofibril / troponin I binding ...pointed-end actin filament capping / lens fiber cell development / myofibril assembly / Striated Muscle Contraction / cytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / myofibril / troponin I binding / filamentous actin / cortical cytoskeleton / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament / skeletal muscle thin filament assembly / actin monomer binding / skeletal muscle fiber development / stress fiber / titin binding / actin filament polymerization / muscle contraction / actin filament organization / sarcomere / adult locomotory behavior / actin filament / filopodium / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / lamellipodium / actin binding / cell body / cytoskeleton / protein domain specific binding / hydrolase activity / calcium ion binding / positive regulation of gene expression / magnesium ion binding / ATP binding / identical protein binding / membrane / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | ||||||||||||
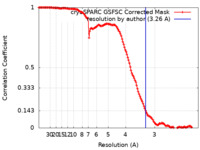
| Method | single particle reconstruction / cryo EM / Resolution: 3.26 Å | ||||||||||||
 Authors Authors | Carman PJ / Barrie KR / Dominguez R | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structures of the free and capped ends of the actin filament. Authors: Peter J Carman / Kyle R Barrie / Grzegorz Rebowski / Roberto Dominguez /  Abstract: The barbed and pointed ends of the actin filament (F-actin) are the sites of growth and shrinkage and the targets of capping proteins that block subunit exchange, including CapZ at the barbed end and ...The barbed and pointed ends of the actin filament (F-actin) are the sites of growth and shrinkage and the targets of capping proteins that block subunit exchange, including CapZ at the barbed end and tropomodulin at the pointed end. We describe cryo-electron microscopy structures of the free and capped ends of F-actin. Terminal subunits at the free barbed end adopt a "flat" F-actin conformation. CapZ binds with minor changes to the barbed end but with major changes to itself. By contrast, subunits at the free pointed end adopt a "twisted" monomeric actin (G-actin) conformation. Tropomodulin binding forces the second subunit into an F-actin conformation. The structures reveal how the ends differ from the middle in F-actin and how these differences control subunit addition, dissociation, capping, and interactions with end-binding proteins. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28936.map.gz emd_28936.map.gz | 123 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28936-v30.xml emd-28936-v30.xml emd-28936.xml emd-28936.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28936_fsc.xml emd_28936_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_28936.png emd_28936.png | 1.4 MB | ||
| Others |  emd_28936_additional_1.map.gz emd_28936_additional_1.map.gz emd_28936_half_map_1.map.gz emd_28936_half_map_1.map.gz emd_28936_half_map_2.map.gz emd_28936_half_map_2.map.gz | 205.7 MB 226.9 MB 226.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28936 http://ftp.pdbj.org/pub/emdb/structures/EMD-28936 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28936 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28936 | HTTPS FTP |
-Related structure data
| Related structure data |  8f8tMC  8f8pC  8f8qC  8f8rC  8f8sC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28936.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28936.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map of the Tropomodulin-bound actin filament pointed end. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharpened final map of the Tropomodulin-bound actin filament...
| File | emd_28936_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened final map of the Tropomodulin-bound actin filament pointed end. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B of the Tropomodulin-bound actin filament pointed end.
| File | emd_28936_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B of the Tropomodulin-bound actin filament pointed end. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map A of the Tropomodulin-bound actin filament pointed end.
| File | emd_28936_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A of the Tropomodulin-bound actin filament pointed end. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tropomodulin-capped pointed end of F-actin
| Entire | Name: Tropomodulin-capped pointed end of F-actin |
|---|---|
| Components |
|
-Supramolecule #1: Tropomodulin-capped pointed end of F-actin
| Supramolecule | Name: Tropomodulin-capped pointed end of F-actin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: Tropomodulin (Tmod)
| Supramolecule | Name: Tropomodulin (Tmod) / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Actin filament
| Supramolecule | Name: Actin filament / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.109973 KDa |
| Sequence | String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIE(HIC)G IIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLD SG DGVTHNVPIY ...String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIE(HIC)G IIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLD SG DGVTHNVPIY EGYALPHAIM RLDLAGRDLT DYLMKILTER GYSFVTTAER EIVRDIKEKL CYVALDFENE MATAASSS S LEKSYELPDG QVITIGNERF RCPETLFQPS FIGMESAGIH ETTYNSIMKC DIDIRKDLYA NNVMSGGTTM YPGIADRMQ KEITALAPST MKIKIIAPPE RKYSVWIGGS ILASLSTFQQ MWITKQEYDE AGPSIVHRKC F |
-Macromolecule #2: Tropomodulin-1
| Macromolecule | Name: Tropomodulin-1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.619078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSYRRELEKY RDLDEDEILG ALTEEELRTL ENELDELDPD NALLPAGLRQ KDQTTKAPTG PFKREELLDH LEKQAKEFKD REDLVPYTG EKRGKVWVPK QKPLDPVLES VTLEPELEEA LANASDAELC DIAAILGMHT LMSNQQYYQA LSSSSIMNKE G LNSVIKPT ...String: MSYRRELEKY RDLDEDEILG ALTEEELRTL ENELDELDPD NALLPAGLRQ KDQTTKAPTG PFKREELLDH LEKQAKEFKD REDLVPYTG EKRGKVWVPK QKPLDPVLES VTLEPELEEA LANASDAELC DIAAILGMHT LMSNQQYYQA LSSSSIMNKE G LNSVIKPT QYKPVPDEEP NSTDVEETLE RIKNNDPKLE EVNLNNIRNI PIPTLKAYAE ALKENSYVKK FSIVGTRSND PV AYALAEM LKENKVLKTL NVESNFISGA GILRLVEALP YNTSLVEMKI DNQSQPLGNK VEMEIVSMLE KNATLLKFGY HFT QQGPRL RASNAMMNNN DLVRKRRLAD LTGPIIPKCR SGV |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 7 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 7 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.05 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force 0 Blot time 2.5 s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 44.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-8f8t: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)