[English] 日本語
 Yorodumi
Yorodumi- EMDB-28876: Thermoplasma acidophilum 20S proteasome - L81Y mutation in alpha ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Thermoplasma acidophilum 20S proteasome - L81Y mutation in alpha subunit | |||||||||
 Map data Map data | L81Y T20S EM Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Protease / threonine protease / endopeptidase activity / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationproteasome endopeptidase complex / proteasome core complex, beta-subunit complex / threonine-type endopeptidase activity / proteasome core complex, alpha-subunit complex / proteasomal protein catabolic process / ubiquitin-dependent protein catabolic process / endopeptidase activity / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) | |||||||||
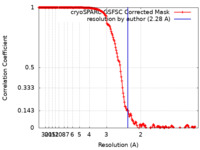
| Method | single particle reconstruction / cryo EM / Resolution: 2.28 Å | |||||||||
 Authors Authors | Chuah J / Smith D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: High resolution structures define divergent and convergent mechanisms of archaeal proteasome activation. Authors: Janelle J Y Chuah / Matthew S Rexroad / David M Smith /  Abstract: Considering the link between neurodegenerative diseases and impaired proteasome function, and the neuro-protective impact of enhanced proteasome activity in animal models, it's crucial to understand ...Considering the link between neurodegenerative diseases and impaired proteasome function, and the neuro-protective impact of enhanced proteasome activity in animal models, it's crucial to understand proteasome activation mechanisms. A hydrophobic-tyrosine-any residue (HbYX) motif on the C-termini of proteasome-activating complexes independently triggers gate-opening of the 20S core particle for protein degradation; however, the causal allosteric mechanism remains unclear. Our study employs a structurally irreducible dipeptide HbYX mimetic to investigate the allosteric mechanism of gate-opening in the archaeal proteasome. High-resolution cryo-EM structures pinpoint vital residues and conformational changes in the proteasome α-subunit implicated in HbYX-dependent activation. Using point mutations, we simulated the HbYX-bound state, providing support for our mechanistic model. We discerned four main mechanistic elements triggering gate-opening: 1) back-loop rearrangement adjacent to K66, 2) intra- and inter- α subunit conformational changes, 3) occupancy of the hydrophobic pocket, and 4) a highly conserved isoleucine-threonine pair in the 20S channel stabilizing the open and closed states, termed the "IT switch." Comparison of different complexes unveiled convergent and divergent mechanism of 20S gate-opening among HbYX-dependent and independent activators. This study delivers a detailed molecular model for HbYX-dependent 20S gate-opening, enabling the development of small molecule proteasome activators that hold promise to treat neurodegenerative diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28876.map.gz emd_28876.map.gz | 449.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28876-v30.xml emd-28876-v30.xml emd-28876.xml emd-28876.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28876_fsc.xml emd_28876_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_28876.png emd_28876.png | 57.2 KB | ||
| Others |  emd_28876_additional_1.map.gz emd_28876_additional_1.map.gz emd_28876_half_map_1.map.gz emd_28876_half_map_1.map.gz emd_28876_half_map_2.map.gz emd_28876_half_map_2.map.gz | 61.6 MB 475.5 MB 475.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28876 http://ftp.pdbj.org/pub/emdb/structures/EMD-28876 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28876 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28876 | HTTPS FTP |
-Validation report
| Summary document |  emd_28876_validation.pdf.gz emd_28876_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28876_full_validation.pdf.gz emd_28876_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_28876_validation.xml.gz emd_28876_validation.xml.gz | 26.6 KB | Display | |
| Data in CIF |  emd_28876_validation.cif.gz emd_28876_validation.cif.gz | 34.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28876 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28876 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28876 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28876 | HTTPS FTP |
-Related structure data
| Related structure data |  8f66MC  8f6aC  8f7kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28876.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28876.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | L81Y T20S EM Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.63281 Å | ||||||||||||||||||||||||||||||||||||
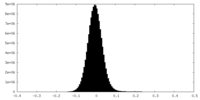
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_28876_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
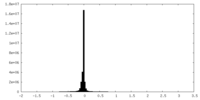
| Density Histograms |
-Half map: L81Y T20S EM Map Half A
| File | emd_28876_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | L81Y T20S EM Map Half A | ||||||||||||
| Projections & Slices |
| ||||||||||||
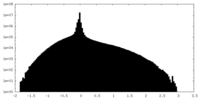
| Density Histograms |
-Half map: L81Y T20S EM Map Half B
| File | emd_28876_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | L81Y T20S EM Map Half B | ||||||||||||
| Projections & Slices |
| ||||||||||||
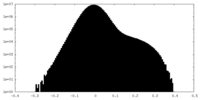
| Density Histograms |
- Sample components
Sample components
-Entire : Thermoplasma acidophilum 20S proteasome - alphaL81Y mutant
| Entire | Name: Thermoplasma acidophilum 20S proteasome - alphaL81Y mutant |
|---|---|
| Components |
|
-Supramolecule #1: Thermoplasma acidophilum 20S proteasome - alphaL81Y mutant
| Supramolecule | Name: Thermoplasma acidophilum 20S proteasome - alphaL81Y mutant type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2, #1 |
|---|---|
| Source (natural) | Organism:   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) |
| Molecular weight | Theoretical: 700 KDa |
-Macromolecule #1: Proteasome subunit alpha
| Macromolecule | Name: Proteasome subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO / EC number: proteasome endopeptidase complex |
|---|---|
| Source (natural) | Organism:   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) |
| Molecular weight | Theoretical: 25.879465 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQQGQMAYDR AITVFSPDGR LFQVEYAREA VKKGSTALGM KFANGVLLIS DKKVRSRLIE QNSIEKIQLI DDYVAAVTSG YVADARVLV DFARISAQQE KVTYGSLVNI ENLVKRVADQ MQQYTQYGGV RPYGVSLIFA GIDQIGPRLF DCDPAGTINE Y KATAIGSG ...String: MQQGQMAYDR AITVFSPDGR LFQVEYAREA VKKGSTALGM KFANGVLLIS DKKVRSRLIE QNSIEKIQLI DDYVAAVTSG YVADARVLV DFARISAQQE KVTYGSLVNI ENLVKRVADQ MQQYTQYGGV RPYGVSLIFA GIDQIGPRLF DCDPAGTINE Y KATAIGSG KDAVVSFLER EYKENLPEKE AVTLGIKALK SSLEEGEELK APEIASITVG NKYRIYDQEE VKKFL UniProtKB: Proteasome subunit alpha |
-Macromolecule #2: Proteasome subunit beta
| Macromolecule | Name: Proteasome subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 14 / Enantiomer: LEVO / EC number: proteasome endopeptidase complex |
|---|---|
| Source (natural) | Organism:   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) |
| Molecular weight | Theoretical: 23.169811 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNQTLETGTT TVGITLKDAV IMATERRVTM ENFIMHKNGK KLFQIDTYTG MTIAGLVGDA QVLVRYMKAE LELYRLQRRV NMPIEAVAT LLSNMLNQVK YMPYMVQLLV GGIDTAPHVF SIDAAGGSVE DIYASTGSGS PFVYGVLESQ YSEKMTVDEG V DLVIRAIS ...String: MNQTLETGTT TVGITLKDAV IMATERRVTM ENFIMHKNGK KLFQIDTYTG MTIAGLVGDA QVLVRYMKAE LELYRLQRRV NMPIEAVAT LLSNMLNQVK YMPYMVQLLV GGIDTAPHVF SIDAAGGSVE DIYASTGSGS PFVYGVLESQ YSEKMTVDEG V DLVIRAIS AAKQRDSASG GMIDVAVITR KDGYVQLPTD QIESRIRKLG LIL UniProtKB: Proteasome subunit beta |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8f66: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)