+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of HLA-A2 bound to MAGEA8 (232-241) peptide | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HLA / MHC / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationantigen processing and presentation of peptide antigen via MHC class I / lumenal side of endoplasmic reticulum membrane / ER to Golgi transport vesicle membrane / MHC class I protein complex / phagocytic vesicle membrane / recycling endosome membrane / early endosome membrane / immune response / Golgi membrane / cell surface / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||
 Authors Authors | Saotome K / Franklin MC | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM. Authors: Kei Saotome / Drew Dudgeon / Kiersten Colotti / Michael J Moore / Jennifer Jones / Yi Zhou / Ashique Rafique / George D Yancopoulos / Andrew J Murphy / John C Lin / William C Olson / Matthew C Franklin /  Abstract: The recognition of antigenic peptide-MHC (pMHC) molecules by T-cell receptors (TCR) initiates the T-cell mediated immune response. Structural characterization is key for understanding the specificity ...The recognition of antigenic peptide-MHC (pMHC) molecules by T-cell receptors (TCR) initiates the T-cell mediated immune response. Structural characterization is key for understanding the specificity of TCR-pMHC interactions and informing the development of therapeutics. Despite the rapid rise of single particle cryoelectron microscopy (cryoEM), x-ray crystallography has remained the preferred method for structure determination of TCR-pMHC complexes. Here, we report cryoEM structures of two distinct full-length α/β TCR-CD3 complexes bound to their pMHC ligand, the cancer-testis antigen HLA-A2/MAGEA4 (230-239). We also determined cryoEM structures of pMHCs containing MAGEA4 (230-239) peptide and the closely related MAGEA8 (232-241) peptide in the absence of TCR, which provided a structural explanation for the MAGEA4 preference displayed by the TCRs. These findings provide insights into the TCR recognition of a clinically relevant cancer antigen and demonstrate the utility of cryoEM for high-resolution structural analysis of TCR-pMHC interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28574.map.gz emd_28574.map.gz | 152.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28574-v30.xml emd-28574-v30.xml emd-28574.xml emd-28574.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28574.png emd_28574.png | 76.8 KB | ||
| Filedesc metadata |  emd-28574.cif.gz emd-28574.cif.gz | 5.5 KB | ||
| Others |  emd_28574_half_map_1.map.gz emd_28574_half_map_1.map.gz emd_28574_half_map_2.map.gz emd_28574_half_map_2.map.gz | 140.9 MB 140.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28574 http://ftp.pdbj.org/pub/emdb/structures/EMD-28574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28574 | HTTPS FTP |
-Related structure data
| Related structure data |  8esbMC  8es7C  8es8C  8es9C  8esaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28574.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28574.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
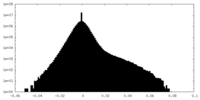
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_28574_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
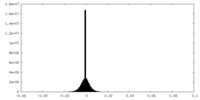
| Density Histograms |
-Half map: #2
| File | emd_28574_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
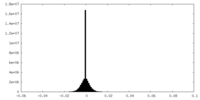
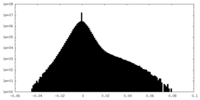
| Density Histograms |
- Sample components
Sample components
-Entire : Single chain disulfide stabilized trimer of HLA-A2, Beta-2-microg...
| Entire | Name: Single chain disulfide stabilized trimer of HLA-A2, Beta-2-microglobulin, MAGE-A8 peptide in complex with anti-Beta-2-microglobulin Fab 2M2 |
|---|---|
| Components |
|
-Supramolecule #1: Single chain disulfide stabilized trimer of HLA-A2, Beta-2-microg...
| Supramolecule | Name: Single chain disulfide stabilized trimer of HLA-A2, Beta-2-microglobulin, MAGE-A8 peptide in complex with anti-Beta-2-microglobulin Fab 2M2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Beta-2-microglobulin,HLA class I antigen,MAGE-A8 peptide chimera
| Macromolecule | Name: Beta-2-microglobulin,HLA class I antigen,MAGE-A8 peptide chimera type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.306184 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GLYDGREHSV GCGGSGGGGS GGGGSIQRTP KIQVYSRHPA ENGKSNFLNC YVSGFHPSDI EVDLLKNGER IEKVEHSDLS FSKDWSFYL LYYTEFTPTE KDEYACRVNH VTLSQPKIVK WDRDMGGGGS GGGGSGGGGS GGGGSGSHSM RYFFTSVSRP G RGEPRFIA ...String: GLYDGREHSV GCGGSGGGGS GGGGSIQRTP KIQVYSRHPA ENGKSNFLNC YVSGFHPSDI EVDLLKNGER IEKVEHSDLS FSKDWSFYL LYYTEFTPTE KDEYACRVNH VTLSQPKIVK WDRDMGGGGS GGGGSGGGGS GGGGSGSHSM RYFFTSVSRP G RGEPRFIA VGYVDDTQFV RFDSDAASQR MEPRAPWIEQ EGPEYWDGET RKVKAHSQTH RVDLGTLRGC YNQSEAGSHT VQ RMYGCDV GSDWRFLRGY HQYAYDGKDY IALKEDLRSW TAADMAAQTT KHKWEAAHVA EQLRAYLEGT CVEWLRRYLE NGK ETLQRT DAPKTHMTHH AVSDHEATLR CWALSFYPAE ITLTWQRDGE DQTQDTELVE TRPAGDGTFQ KWAAVVVPSG QEQR YTCHV QHEGLPKPLT LRWEPEQKLI SEEDLGGEQK LISEEDLHHH HHH UniProtKB: HLA class I antigen |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.12 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 121731 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)