+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focused reconstruction of HRP29 tail | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | HRP29 / VIRUS | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont genome ejection through host cell envelope, short tail mechanism / virus tail / outer membrane / virion component Similarity search - Function | |||||||||||||||
| Biological species |  Shigella phage Buco (virus) Shigella phage Buco (virus) | |||||||||||||||
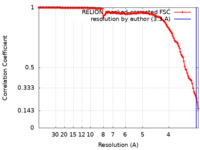
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||
 Authors Authors | Subramanian S / Bergland Drarvik SM / Parent KN | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of Shigella bacteriophage HRP29 Authors: Subramanian S / Bergland Drarvik SM / Parent KN | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28562.map.gz emd_28562.map.gz | 8.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28562-v30.xml emd-28562-v30.xml emd-28562.xml emd-28562.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28562_fsc.xml emd_28562_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_28562.png emd_28562.png | 150.8 KB | ||
| Masks |  emd_28562_msk_1.map emd_28562_msk_1.map | 209.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28562.cif.gz emd-28562.cif.gz | 6 KB | ||
| Others |  emd_28562_half_map_1.map.gz emd_28562_half_map_1.map.gz emd_28562_half_map_2.map.gz emd_28562_half_map_2.map.gz | 163.3 MB 163.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28562 http://ftp.pdbj.org/pub/emdb/structures/EMD-28562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28562 | HTTPS FTP |
-Related structure data
| Related structure data |  8es4MC  8eldC  8em6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28562.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28562.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.632 Å | ||||||||||||||||||||||||||||||||||||
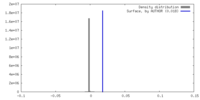
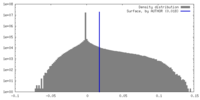
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28562_msk_1.map emd_28562_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
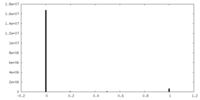
| Density Histograms |
-Half map: #1
| File | emd_28562_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
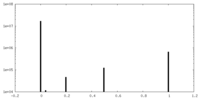
| Density Histograms |
-Half map: #2
| File | emd_28562_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Shigella phage Buco
| Entire | Name:  Shigella phage Buco (virus) Shigella phage Buco (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Shigella phage Buco
| Supramolecule | Name: Shigella phage Buco / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2530183 / Sci species name: Shigella phage Buco / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Gp35
| Macromolecule | Name: Gp35 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella phage Buco (virus) Shigella phage Buco (virus) |
| Molecular weight | Theoretical: 56.983391 KDa |
| Sequence | String: MQGQQKESLE SLFTKDSDPT VLDAAEQFAQ WTLPTVLTRD ISGMDGKRTS LHRDYQSTGA VLVNSASTKV TNALFPQGAP FFRFVDSPD MAAAVAELGI NGTVQSQQSQ IELSASSLVF SRDNYAASLR AVKLLMVTGN ALEYFDEGTG RSHIYSVREY T VRRDGSGN ...String: MQGQQKESLE SLFTKDSDPT VLDAAEQFAQ WTLPTVLTRD ISGMDGKRTS LHRDYQSTGA VLVNSASTKV TNALFPQGAP FFRFVDSPD MAAAVAELGI NGTVQSQQSQ IELSASSLVF SRDNYAASLR AVKLLMVTGN ALEYFDEGTG RSHIYSVREY T VRRDGSGN ILRVVLKERI AAMDLPQEFR SAHLGQKDDY DDVTLYTGIC LEDNKFKIYQ EVQQQQIGDA STYPIDECPY TV LVWNLVN GEHYGRGLVE DYAGDFARLS VLSQALTLYE VEAARLYNAV SAGAGIDVDA AQAAETGDYV QTSAAPGTNP GIW AVENGS DRKIMSLQSE ISMIEQKLAR AFMYAGNTRQ GERVTAYEIR TNAQEAQNSL GDAYSILSDH WLRKRAYLYT VYQY PPMRA MFTLGATTIQ ILVGTASLNK AAQADRLLEA SQSIQLVLPV LQGATKRTNP DAVVDFILDA FGVVSSKLMY TEEQL KQIQ DQQDQQQADQ QRNLELAQAN PEVAGQQLGL IPS UniProtKB: Head-tail connector protein |
-Macromolecule #2: Gp39
| Macromolecule | Name: Gp39 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella phage Buco (virus) Shigella phage Buco (virus) |
| Molecular weight | Theoretical: 20.81551 KDa |
| Sequence | String: MRLTDAVNVT LEALGESRIV DINTSNPSAG LARAALDRTR RGVLSTGWWF NTIIREVTPT PNPGQIKVPW NQLSMYGLDG TKYGERDGV LYNLVDQTKV FSDTVHLKVV IDIDFEDLPE HMAMWVANAT AAQVYLNDLG ADGNYKSLLG IAAEYEAMNM R EHLRNQRY STSRTHAARK IRSGFFR UniProtKB: Putative tail protein |
-Macromolecule #3: Gp40
| Macromolecule | Name: Gp40 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella phage Buco (virus) Shigella phage Buco (virus) |
| Molecular weight | Theoretical: 84.783617 KDa |
| Sequence | String: MAQTFEGTLQ SLLQGVSQQI PRERQPGQLG AQQNMLSDPV TGLRRRPPLH LAAQTLMENP VSPDALFSTY IERGTDGRHL LINTEAGIW QILSKDATTL IRSGQADYLK ASIGATSIQT ASIAGLTYIL NTEQTPVAHV DNTGKLNPAN TGFFYIVASS F SKRWTITV ...String: MAQTFEGTLQ SLLQGVSQQI PRERQPGQLG AQQNMLSDPV TGLRRRPPLH LAAQTLMENP VSPDALFSTY IERGTDGRHL LINTEAGIW QILSKDATTL IRSGQADYLK ASIGATSIQT ASIAGLTYIL NTEQTPVAHV DNTGKLNPAN TGFFYIVASS F SKRWTITV QSNEGTWTAV HDVGASSDDG AVPAATASAV INSLKTNLLA AGMPSDKVDT FGSYMFIKGL TNVVVSSDAG TT YARWSNQ SRVDEESDLP AQLPASANGC MCRVGAASTS ATWYRFDYAT RQWNEDSAYS SITKITNMPL EFAADDQIIP RDF EGRLAG DDENNEDPGF VENGYITGIA AFQGRLVLLS GSRVSMSASG LYQRFYRSTV VNLLDTDRID IGAASAQDSV FRAA LQFNR DLVVFGDSMQ AVIAGNAVLT PTNASIALTS EFSCDSRVIP VVTGQTVLYA SRRNSDYAGL LEFIPSAYTS SQYVS QDAT VHLPRYIPGR VMDMQVSSVT NVAFFRYSGE RTSVLVYEFL WGEDAKRAQG AYHKWVLPYD VLSLHTLSEA AYFFVR GPG AYVLALRVDP REGFVAGTTY EYPFMDMGAP VTVQGGQFTL PEHLRKAGLQ DSIALAYYTG DDSGSELGIA SISSNWV CT TVRGVPDGNY LAGYRFKSGT TLTPPMLKDQ NDNLIGSGHV RLLRLDVAMR NSGVVDVLVE DNARDVDNDS EYSGVLMN S KELAPEQPLK ASLSNIIIPC RTNTDTTEVT LSTSGTLEMN IMDVSYILRY NQRRRRV UniProtKB: Putative tail protein |
-Macromolecule #4: Gp44
| Macromolecule | Name: Gp44 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella phage Buco (virus) Shigella phage Buco (virus) |
| Molecular weight | Theoretical: 28.595883 KDa |
| Sequence | String: MAYSWSEQVV PSGTTLISVD IEYLDKSYIY LYINNVLISN SDYSWNSDTL IQLNTPMASA GTVLLVRRTD KEYLYIMFAE GAAFIRENL DVQNTQFLHL AQELVEGRSI DGFYGDLSMN GYRITHLADG VDPKDAVNKG QLDSVSNRVS SIENSFLGLT T VSYPWYTV ...String: MAYSWSEQVV PSGTTLISVD IEYLDKSYIY LYINNVLISN SDYSWNSDTL IQLNTPMASA GTVLLVRRTD KEYLYIMFAE GAAFIRENL DVQNTQFLHL AQELVEGRSI DGFYGDLSMN GYRITHLADG VDPKDAVNKG QLDSVSNRVS SIENSFLGLT T VSYPWYTV VSADTDTFEP PFKFTKAALY IDGLCQVPDY SYVVVDNKLL LAESVPTGTV VFARLGEDTD AATEAATTTA LA AVQADLQ NQINALRALL QGG UniProtKB: Putative tail protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 33.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)