[English] 日本語
 Yorodumi
Yorodumi- EMDB-2846: Three-dimensional structure of the autophagic phosphatidylinosito... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2846 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
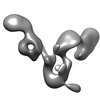
| Title | Three-dimensional structure of the autophagic phosphatidylinositol 3-kinase complex. | |||||||||
 Map data Map data | Three-dimensional structure of the autophagic phosphatidylinositol 3-kinase complex, PI3KC3-C1. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | kinase / lipid kinase / protein kinase / autophagy / phosphatidylinositol 3-kinase / PI3K / phosphatidylinositol 3-phosphate / PI(3)P | |||||||||
| Function / homology |  Function and homology information Function and homology informationextrinsic component of omegasome membrane / phosphatidylinositol 3-kinase inhibitor activity / regulation of triglyceride metabolic process / extrinsic component of phagophore assembly site membrane / nucleus-vacuole junction / cellular response to aluminum ion / positive regulation of protein lipidation / postsynaptic endosome / Toll Like Receptor 9 (TLR9) Cascade / Synthesis of PIPs at the late endosome membrane ...extrinsic component of omegasome membrane / phosphatidylinositol 3-kinase inhibitor activity / regulation of triglyceride metabolic process / extrinsic component of phagophore assembly site membrane / nucleus-vacuole junction / cellular response to aluminum ion / positive regulation of protein lipidation / postsynaptic endosome / Toll Like Receptor 9 (TLR9) Cascade / Synthesis of PIPs at the late endosome membrane / positive regulation of stress granule assembly / phosphatidylinositol 3-kinase complex, class III / cellular response to oxygen-glucose deprivation / Synthesis of PIPs at the early endosome membrane / phosphatidylinositol 3-kinase complex, class III, type II / phosphatidylinositol 3-kinase complex, class III, type I / host-mediated activation of viral genome replication / response to mitochondrial depolarisation / presynaptic endosome / positive regulation of attachment of mitotic spindle microtubules to kinetochore / mitochondria-associated endoplasmic reticulum membrane contact site / negative regulation of lysosome organization / engulfment of apoptotic cell / Synthesis of PIPs at the Golgi membrane / phosphatidylinositol kinase activity / regulation of protein complex stability / positive regulation of autophagosome assembly / early endosome to late endosome transport / response to L-leucine / cytoplasmic side of mitochondrial outer membrane / negative regulation of autophagosome assembly / protein localization to phagophore assembly site / phosphatidylinositol 3-kinase regulator activity / receptor catabolic process / phagophore assembly site membrane / protein targeting to vacuole / protein targeting to lysosome / SMAD protein signal transduction / late endosome to vacuole transport / endosome organization / pexophagy / phagophore assembly site / Translation of Replicase and Assembly of the Replication Transcription Complex / phosphatidylinositol-3-phosphate biosynthetic process / cellular response to nitrogen starvation / negative regulation of programmed cell death / phosphatidylinositol 3-kinase / 1-phosphatidylinositol-3-kinase activity / lysosome organization / response to vitamin E / post-transcriptional regulation of gene expression / mitotic metaphase chromosome alignment / response to iron(II) ion / cytoplasmic pattern recognition receptor signaling pathway / endosome to lysosome transport / positive regulation of cardiac muscle hypertrophy / Macroautophagy / phosphatidylinositol-mediated signaling / autophagosome membrane docking / RSV-host interactions / p38MAPK cascade / phosphatidylinositol phosphate biosynthetic process / negative regulation of protein phosphorylation / autolysosome / autophagosome membrane / PI3K Cascade / axoneme / synaptic vesicle endocytosis / autophagosome assembly / RHO GTPases Activate NADPH Oxidases / amyloid-beta metabolic process / autophagosome maturation / regulation of macroautophagy / cellular defense response / neuron development / cellular response to glucose starvation / phosphatidylinositol 3-kinase binding / mitophagy / intercellular bridge / positive regulation of intrinsic apoptotic signaling pathway / phagocytic vesicle / JNK cascade / protein-membrane adaptor activity / positive regulation of autophagy / cellular response to amino acid starvation / autophagosome / cellular response to copper ion / cellular response to epidermal growth factor stimulus / cellular response to starvation / regulation of cytokinesis / Antigen Presentation: Folding, assembly and peptide loading of class I MHC / macroautophagy / phosphatidylinositol 3-kinase/protein kinase B signal transduction / trans-Golgi network / circadian rhythm / GABA-ergic synapse / protein processing / response to lead ion / ISG15 antiviral mechanism / autophagy Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 27.5 Å | |||||||||
 Authors Authors | Baskaran S / Carlson L-A / Stjepanovic G / Young LN / Kim DJ / Grob P / Stanley RE / Nogales E / Hurley JH | |||||||||
 Citation Citation |  Journal: Elife / Year: 2014 Journal: Elife / Year: 2014Title: Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Authors: Sulochanadevi Baskaran / Lars-Anders Carlson / Goran Stjepanovic / Lindsey N Young / Do Jin Kim / Patricia Grob / Robin E Stanley / Eva Nogales / James H Hurley /  Abstract: The class III phosphatidylinositol 3-kinase complex I (PI3KC3-C1) that functions in early autophagy consists of the lipid kinase VPS34, the scaffolding protein VPS15, the tumor suppressor BECN1, and ...The class III phosphatidylinositol 3-kinase complex I (PI3KC3-C1) that functions in early autophagy consists of the lipid kinase VPS34, the scaffolding protein VPS15, the tumor suppressor BECN1, and the autophagy-specific subunit ATG14. The structure of the ATG14-containing PI3KC3-C1 was determined by single-particle EM, revealing a V-shaped architecture. All of the ordered domains of VPS34, VPS15, and BECN1 were mapped by MBP tagging. The dynamics of the complex were defined using hydrogen-deuterium exchange, revealing a novel 20-residue ordered region C-terminal to the VPS34 C2 domain. VPS15 organizes the complex and serves as a bridge between VPS34 and the ATG14:BECN1 subcomplex. Dynamic transitions occur in which the lipid kinase domain is ejected from the complex and VPS15 pivots at the base of the V. The N-terminus of BECN1, the target for signaling inputs, resides near the pivot point. These observations provide a framework for understanding the allosteric regulation of lipid kinase activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2846.map.gz emd_2846.map.gz | 3.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2846-v30.xml emd-2846-v30.xml emd-2846.xml emd-2846.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2846.tif emd_2846.tif | 86.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2846 http://ftp.pdbj.org/pub/emdb/structures/EMD-2846 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2846 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2846 | HTTPS FTP |
-Validation report
| Summary document |  emd_2846_validation.pdf.gz emd_2846_validation.pdf.gz | 199 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2846_full_validation.pdf.gz emd_2846_full_validation.pdf.gz | 198.1 KB | Display | |
| Data in XML |  emd_2846_validation.xml.gz emd_2846_validation.xml.gz | 5.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2846 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2846 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2846 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2846 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2846.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2846.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Three-dimensional structure of the autophagic phosphatidylinositol 3-kinase complex, PI3KC3-C1. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.01 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Autophagic phosphatidylinositol 3-kinase complex, PI3KC3-C1.
| Entire | Name: Autophagic phosphatidylinositol 3-kinase complex, PI3KC3-C1. |
|---|---|
| Components |
|
-Supramolecule #1000: Autophagic phosphatidylinositol 3-kinase complex, PI3KC3-C1.
| Supramolecule | Name: Autophagic phosphatidylinositol 3-kinase complex, PI3KC3-C1. type: sample / ID: 1000 / Oligomeric state: heterotetramer / Number unique components: 4 |
|---|---|
| Molecular weight | Theoretical: 361.8 KDa |
-Macromolecule #1: phosphatidylinositol 3-kinase, catalytic subunit type 3
| Macromolecule | Name: phosphatidylinositol 3-kinase, catalytic subunit type 3 type: protein_or_peptide / ID: 1 / Name.synonym: VPS34, PI3KC3 / Number of copies: 1 / Oligomeric state: one component of heterotetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Organelle: autophagosome / Location in cell: autophagosome membrane Homo sapiens (human) / synonym: Human / Organelle: autophagosome / Location in cell: autophagosome membrane |
| Molecular weight | Theoretical: 102 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 / Recombinant plasmid: pCAG Homo sapiens (human) / Recombinant cell: HEK293 / Recombinant plasmid: pCAG |
| Sequence | UniProtKB: Phosphatidylinositol 3-kinase catalytic subunit type 3 GO: 1-phosphatidylinositol-3-kinase activity, phosphatidylinositol 3-kinase complex, class III, type I, autophagy, autophagosome assembly |
-Macromolecule #2: phosphoinositide-3-kinase, regulatory subunit 4
| Macromolecule | Name: phosphoinositide-3-kinase, regulatory subunit 4 / type: protein_or_peptide / ID: 2 / Name.synonym: VPS15, p150, PIK3R4 / Number of copies: 1 / Oligomeric state: one component of heterotetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Organelle: autophagosome / Location in cell: autophagosome membrane Homo sapiens (human) / synonym: Human / Organelle: autophagosome / Location in cell: autophagosome membrane |
| Molecular weight | Theoretical: 153 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 / Recombinant plasmid: pCAG Homo sapiens (human) / Recombinant cell: HEK293 / Recombinant plasmid: pCAG |
| Sequence | UniProtKB: Phosphoinositide 3-kinase regulatory subunit 4 GO: phosphatidylinositol 3-kinase complex, class III, type I, autophagy, autophagosome assembly |
-Macromolecule #3: beclin1
| Macromolecule | Name: beclin1 / type: protein_or_peptide / ID: 3 / Name.synonym: BECN1 / Number of copies: 1 / Oligomeric state: one component of heterotetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Organelle: autophagosome / Location in cell: autophagosome membrane Homo sapiens (human) / synonym: Human / Organelle: autophagosome / Location in cell: autophagosome membrane |
| Molecular weight | Theoretical: 52 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 / Recombinant plasmid: pCAG Homo sapiens (human) / Recombinant cell: HEK293 / Recombinant plasmid: pCAG |
| Sequence | UniProtKB: Beclin-1 GO: phosphatidylinositol 3-kinase complex, class III, type I, autophagy, autophagosome assembly |
-Macromolecule #4: autophagy related 14
| Macromolecule | Name: autophagy related 14 / type: protein_or_peptide / ID: 4 / Name.synonym: ATG14, ATG14L, BARKOR / Number of copies: 1 / Oligomeric state: one component of heterotetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Organelle: autophagosome / Location in cell: autophagosome membrane Homo sapiens (human) / synonym: Human / Organelle: autophagosome / Location in cell: autophagosome membrane |
| Molecular weight | Theoretical: 55 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 / Recombinant plasmid: pCAG Homo sapiens (human) / Recombinant cell: HEK293 / Recombinant plasmid: pCAG |
| Sequence | UniProtKB: Beclin 1-associated autophagy-related key regulator GO: phosphatidylinositol 3-kinase complex, class III, type I, autophagy, autophagosome assembly |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM Tris-HCl, 200 mM NaCl, 2 mM MgCl2, 1 mM TCEP, 3% trehalose |
| Staining | Type: NEGATIVE Details: Protein was incubated on grids for 30s, followed by sequential 10 s incubations on four 50 ul drops of 1% uranyl formate. The stained grids were blotted to near dryness with a filter paper and air-dried. |
| Grid | Details: Continuous carbon, plasma cleaned for 10 s in a Gatan Solarus at 10% O2. |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 292 K / Max: 294 K / Average: 293 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 80,000 times magnification. |
| Date | May 28, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 1614 / Average electron dose: 25 e/Å2 / Bits/pixel: 32 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 100333 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 45 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The final three-dimensional structure was calculated in RELION using a single 3D class. Starting model for this calculation was a random conical tilt model low-pass filtered to 80A. |
|---|---|
| CTF correction | Details: tilt-dependent phase flip |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 27.5 Å / Resolution method: OTHER / Software - Name: Spider, EMAN, EMAN2, IMAGIC, RELION Details: Final map calculated in RELION using a single 3D class. Data acquired at 0, 30, and 45 degrees were used to refine an initial RCT model low-pass filtered to 80A. Gold standard FSC calculated using RELION. Number images used: 38745 |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)