[English] 日本語
 Yorodumi
Yorodumi- EMDB-28268: Phospholipase C beta 3 (PLCb3) in complex with Gbg on lipid nanodiscs -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Phospholipase C beta 3 (PLCb3) in complex with Gbg on lipid nanodiscs | |||||||||
 Map data Map data | final sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PIP2 degradation / IP3 production / DAG production / G protein signaling / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphoinositide phospholipase C / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phosphatidylinositol metabolic process / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / regulation of systemic arterial blood pressure / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / C-type glycerophospholipase activity / phosphatidylinositol-mediated signaling ...phosphoinositide phospholipase C / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phosphatidylinositol metabolic process / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / regulation of systemic arterial blood pressure / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / C-type glycerophospholipase activity / phosphatidylinositol-mediated signaling / Synthesis of IP3 and IP4 in the cytosol / postsynaptic cytosol / lipid catabolic process / release of sequestered calcium ion into cytosol / molecular function activator activity / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / molecular adaptor activity / Ras protein signal transduction / calmodulin binding / Extra-nuclear estrogen signaling / cell population proliferation / cadherin binding / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / calcium ion binding / synapse / protein-containing complex binding / signal transduction / protein-containing complex / extracellular exosome / membrane / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
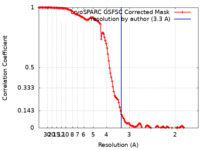
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Falzone ME / MacKinnon R | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: activates hydrolysis by recruiting and orienting on the membrane surface. Authors: Maria E Falzone / Roderick MacKinnon /  Abstract: catalyze the hydrolysis of phosphatidylinositol 4, 5-bisphosphate [Formula: see text] into [Formula: see text] [Formula: see text] and [Formula: see text] [Formula: see text]. [Formula: see text] ... catalyze the hydrolysis of phosphatidylinositol 4, 5-bisphosphate [Formula: see text] into [Formula: see text] [Formula: see text] and [Formula: see text] [Formula: see text]. [Formula: see text] regulates the activity of many membrane proteins, while and lead to increased intracellular Ca levels and activate protein kinase C, respectively. are regulated by G protein-coupled receptors through direct interaction with [Formula: see text] and [Formula: see text] and are aqueous-soluble enzymes that must bind to the cell membrane to act on their lipid substrate. This study addresses the mechanism by which [Formula: see text] activates 3. We show that 3 functions as a slow Michaelis-Menten enzyme ( [Formula: see text] ) on membrane surfaces. We used membrane partitioning experiments to study the solution-membrane localization equilibrium of 3. Its partition coefficient is such that only a small quantity of 3 exists in the membrane in the absence of [Formula: see text] . When [Formula: see text] is present, equilibrium binding on the membrane surface increases 3 in the membrane, increasing [Formula: see text] in proportion. Atomic structures on membrane vesicle surfaces show that two [Formula: see text] anchor 3 with its catalytic site oriented toward the membrane surface. Taken together, the enzyme kinetic, membrane partitioning, and structural data show that [Formula: see text] activates by increasing its concentration on the membrane surface and orienting its catalytic core to engage [Formula: see text] . This principle of activation explains rapid stimulated catalysis with low background activity, which is essential to the biological processes mediated by [Formula: see text], and . | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28268.map.gz emd_28268.map.gz | 204 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28268-v30.xml emd-28268-v30.xml emd-28268.xml emd-28268.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28268_fsc.xml emd_28268_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_28268.png emd_28268.png | 41.4 KB | ||
| Filedesc metadata |  emd-28268.cif.gz emd-28268.cif.gz | 6.9 KB | ||
| Others |  emd_28268_additional_1.map.gz emd_28268_additional_1.map.gz emd_28268_half_map_1.map.gz emd_28268_half_map_1.map.gz emd_28268_half_map_2.map.gz emd_28268_half_map_2.map.gz | 108.2 MB 200.6 MB 200.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28268 http://ftp.pdbj.org/pub/emdb/structures/EMD-28268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28268 | HTTPS FTP |
-Related structure data
| Related structure data |  8emxMC  8emvC  8emwC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28268.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28268.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | final sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.844 Å | ||||||||||||||||||||||||||||||||||||
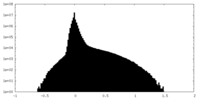
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: final unsharpened map
| File | emd_28268_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | final unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
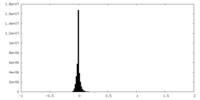
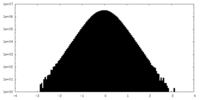
| Density Histograms |
-Half map: #1
| File | emd_28268_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
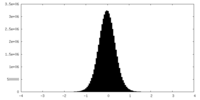
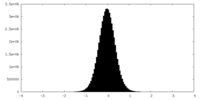
| Density Histograms |
-Half map: #2
| File | emd_28268_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
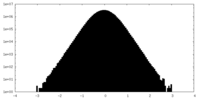
| Density Histograms |
- Sample components
Sample components
+Entire : Phospholipase C beta 3 (PLCb3) in complex with two copies of Gbg ...
+Supramolecule #1: Phospholipase C beta 3 (PLCb3) in complex with two copies of Gbg ...
+Supramolecule #2: Phospholipase C beta 3 (PLCb3)
+Supramolecule #3: G protein beta subunit (Gb1)
+Supramolecule #4: G protein gamma subunit 2 (Gg2)
+Supramolecule #5: G protein beta subunit (Gb1)
+Supramolecule #6: G protein gamma subunit 2 (Gg2)
+Macromolecule #1: 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3
+Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
+Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
+Macromolecule #4: CALCIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 22 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 2.0 sec. / Average electron dose: 69.14 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






































 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)