[English] 日本語
 Yorodumi
Yorodumi- EMDB-28053: Local refined map of the TMD region of Human GluN1a-GluN2D NMDA r... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Local refined map of the TMD region of Human GluN1a-GluN2D NMDA receptor | |||||||||
 Map data Map data | TMD local refined map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.92 Å | |||||||||
 Authors Authors | Kang H / Furukawa H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs. Authors: Tsung-Han Chou / Hyunook Kang / Noriko Simorowski / Stephen F Traynelis / Hiro Furukawa /  Abstract: Neurotransmission mediated by diverse subtypes of N-methyl-D-aspartate receptors (NMDARs) is fundamental for basic brain functions and development as well as neuropsychiatric diseases and disorders. ...Neurotransmission mediated by diverse subtypes of N-methyl-D-aspartate receptors (NMDARs) is fundamental for basic brain functions and development as well as neuropsychiatric diseases and disorders. NMDARs are glycine- and glutamate-gated ion channels that exist as heterotetramers composed of obligatory GluN1 and GluN2(A-D) and/or GluN3(A-B). The GluN2C and GluN2D subunits form ion channels with distinct properties and spatio-temporal expression patterns. Here, we provide the structures of the agonist-bound human GluN1-2C NMDAR in the presence and absence of the GluN2C-selective positive allosteric potentiator (PAM), PYD-106, the agonist-bound GluN1-2A-2C tri-heteromeric NMDAR, and agonist-bound GluN1-2D NMDARs by single-particle electron cryomicroscopy. Our analysis shows unique inter-subunit and domain arrangements of the GluN2C NMDARs, which contribute to functional regulation and formation of the PAM binding pocket and is distinct from GluN2D NMDARs. Our findings here provide the fundamental blueprint to study GluN2C- and GluN2D-containing NMDARs, which are uniquely involved in neuropsychiatric disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28053.map.gz emd_28053.map.gz | 229.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28053-v30.xml emd-28053-v30.xml emd-28053.xml emd-28053.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28053.png emd_28053.png | 37.6 KB | ||
| Others |  emd_28053_half_map_1.map.gz emd_28053_half_map_1.map.gz emd_28053_half_map_2.map.gz emd_28053_half_map_2.map.gz | 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28053 http://ftp.pdbj.org/pub/emdb/structures/EMD-28053 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28053 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28053 | HTTPS FTP |
-Validation report
| Summary document |  emd_28053_validation.pdf.gz emd_28053_validation.pdf.gz | 951.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28053_full_validation.pdf.gz emd_28053_full_validation.pdf.gz | 950.8 KB | Display | |
| Data in XML |  emd_28053_validation.xml.gz emd_28053_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_28053_validation.cif.gz emd_28053_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28053 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28053 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28053 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28053 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28053.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28053.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TMD local refined map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
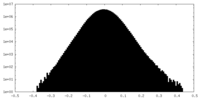
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_28053_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
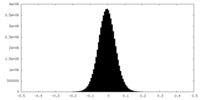
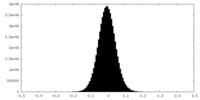
| Density Histograms |
-Half map: #2
| File | emd_28053_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
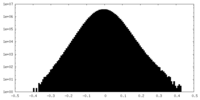
| Density Histograms |
- Sample components
Sample components
-Entire : Hetero-tetrameric GluN1a-GluN2D NMDA receptors
| Entire | Name: Hetero-tetrameric GluN1a-GluN2D NMDA receptors |
|---|---|
| Components |
|
-Supramolecule #1: Hetero-tetrameric GluN1a-GluN2D NMDA receptors
| Supramolecule | Name: Hetero-tetrameric GluN1a-GluN2D NMDA receptors / type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 285 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 74.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)