+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human ABCA7 in BPL/Ch nanodiscs (Map 2) | |||||||||
 Map data Map data | human ABCA7 in BPL/Ch nanodiscs, alternative 3D class Map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Alam A / Le LTM / Thompson JR | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms. Authors: Le Thi My Le / James Robert Thompson / Sepehr Dehghani-Ghahnaviyeh / Shashank Pant / Phuoc Xuan Dang / Jarrod Bradley French / Takahisa Kanikeyo / Emad Tajkhorshid / Amer Alam /  Abstract: Phospholipid extrusion by ABC subfamily A (ABCA) exporters is central to cellular physiology, although the specifics of the underlying substrate interactions and transport mechanisms remain poorly ...Phospholipid extrusion by ABC subfamily A (ABCA) exporters is central to cellular physiology, although the specifics of the underlying substrate interactions and transport mechanisms remain poorly resolved at the molecular level. Here we report cryo-EM structures of lipid-embedded human ABCA7 in an open state and in a nucleotide-bound, closed state at resolutions between 3.6 and 4.0 Å. The former reveals an ordered patch of bilayer lipids traversing the transmembrane domain (TMD), while the latter reveals a lipid-free, closed TMD with a small extracellular opening. These structures offer a structural framework for both substrate entry and exit from the ABCA7 TMD and highlight conserved rigid-body motions that underlie the associated conformational transitions. Combined with functional analysis and molecular dynamics (MD) simulations, our data also shed light on lipid partitioning into the ABCA7 TMD and localized membrane perturbations that underlie ABCA7 function and have broader implications for other ABCA family transporters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28044.map.gz emd_28044.map.gz | 202.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28044-v30.xml emd-28044-v30.xml emd-28044.xml emd-28044.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28044.png emd_28044.png | 26.7 KB | ||
| Others |  emd_28044_half_map_1.map.gz emd_28044_half_map_1.map.gz emd_28044_half_map_2.map.gz emd_28044_half_map_2.map.gz | 171.2 MB 171.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28044 http://ftp.pdbj.org/pub/emdb/structures/EMD-28044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28044 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28044.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28044.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human ABCA7 in BPL/Ch nanodiscs, alternative 3D class Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.895 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Refinement half map1
| File | emd_28044_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
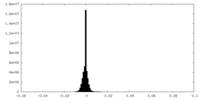
| Density Histograms |
-Half map: Refinement half map2
| File | emd_28044_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
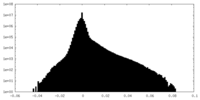
| Density Histograms |
- Sample components
Sample components
-Entire : Human ABCA7 in Porcine BPL/Ch/MSP1D1 nanodiscs
| Entire | Name: Human ABCA7 in Porcine BPL/Ch/MSP1D1 nanodiscs |
|---|---|
| Components |
|
-Supramolecule #1: Human ABCA7 in Porcine BPL/Ch/MSP1D1 nanodiscs
| Supramolecule | Name: Human ABCA7 in Porcine BPL/Ch/MSP1D1 nanodiscs / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 Details: Human ABCA7 recombinantly expressed in HEK293 TREX cell line and reconstituted in MSP1D1 nanodiscs comprising 4:1 mixture of Porcine Brain Polar Lipids:Cholesterol |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 234.35 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 25mM HEPES pH 7.5, 150mM NaCl, 5mM ATP gammaS, 10mM Magnesium Chloride |
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average exposure time: 60.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)