[English] 日本語
 Yorodumi
Yorodumi- EMDB-28033: cryo-EM structure of TRPM3 ion channel in the presence with PIP2 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of TRPM3 ion channel in the presence with PIP2 and PregS, state 2 | |||||||||
 Map data Map data | TRPM3 in the presence of PIP2 and PregS, state 2, sharpened by a global B-factor of 121.5 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRPM3 / ion channel / PIP2 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationzinc ion transmembrane transporter activity / zinc ion transmembrane transport / temperature-gated ion channel activity / sodium ion transport / monoatomic cation transport / monoatomic cation channel activity / phosphatidylinositol-4,5-bisphosphate binding / calcium ion transmembrane transport / calcium channel activity / G-protein beta/gamma-subunit complex binding ...zinc ion transmembrane transporter activity / zinc ion transmembrane transport / temperature-gated ion channel activity / sodium ion transport / monoatomic cation transport / monoatomic cation channel activity / phosphatidylinositol-4,5-bisphosphate binding / calcium ion transmembrane transport / calcium channel activity / G-protein beta/gamma-subunit complex binding / protein homotetramerization / calmodulin binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
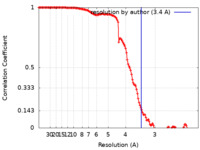
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Zhao C / MacKinnon R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Neuron / Year: 2023 Journal: Neuron / Year: 2023Title: Structural and functional analyses of a GPCR-inhibited ion channel TRPM3. Authors: Chen Zhao / Roderick MacKinnon /  Abstract: G-protein coupled receptors (GPCRs) govern the physiological response to stimuli by modulating the activity of downstream effectors, including ion channels. TRPM3 is an ion channel inhibited by GPCRs ...G-protein coupled receptors (GPCRs) govern the physiological response to stimuli by modulating the activity of downstream effectors, including ion channels. TRPM3 is an ion channel inhibited by GPCRs through direct interaction with G protein (Gβγ) released upon their activation. This GPCR-TRPM3 signaling pathway contributes to the analgesic effect of morphine. Here, we characterized Gβγ inhibition of TRPM3 using electrophysiology and single particle cryo-electron microscopy (cryo-EM). From electrophysiology, we obtained a half inhibition constant (IC50) of ∼240 nM. Using cryo-EM, we determined structures of mouse TRPM3 expressed in human cells with and without Gβγ and with and without PIP, a lipid required for TRPM3 activity, at resolutions of 2.7-4.7 Å. Gβγ-TRPM3 interfaces vary depending on PIP occupancy; however, in all cases, Gβγ appears loosely attached to TRPM3. The IC50 in electrophysiology experiments raises the possibility that additional unknown factors may stabilize the TRPM3-Gβγ complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28033.map.gz emd_28033.map.gz | 203.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28033-v30.xml emd-28033-v30.xml emd-28033.xml emd-28033.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28033_fsc.xml emd_28033_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_28033.png emd_28033.png | 201.7 KB | ||
| Filedesc metadata |  emd-28033.cif.gz emd-28033.cif.gz | 7.2 KB | ||
| Others |  emd_28033_half_map_1.map.gz emd_28033_half_map_1.map.gz emd_28033_half_map_2.map.gz emd_28033_half_map_2.map.gz | 199.8 MB 199.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28033 http://ftp.pdbj.org/pub/emdb/structures/EMD-28033 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28033 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28033 | HTTPS FTP |
-Related structure data
| Related structure data |  8ed9MC  8ddqC  8ddrC  8ddsC  8ddtC  8dduC  8ddvC  8ddwC  8ddxC  8ed7C  8ed8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28033.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28033.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRPM3 in the presence of PIP2 and PregS, state 2, sharpened by a global B-factor of 121.5 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: TRPM3 in the presence of PIP2 and PregS, state 2, half-map 2
| File | emd_28033_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRPM3 in the presence of PIP2 and PregS, state 2, half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: TRPM3 in the presence of PIP2 and PregS, state 2, half-map 1
| File | emd_28033_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRPM3 in the presence of PIP2 and PregS, state 2, half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TRPM3
| Entire | Name: TRPM3 |
|---|---|
| Components |
|
-Supramolecule #1: TRPM3
| Supramolecule | Name: TRPM3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel, subfamily M, member 3
| Macromolecule | Name: Transient receptor potential cation channel, subfamily M, member 3 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 154.649328 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GKKWRDAGEL ERGCSDREDS AESRRRSRSA SRGRFAESWK RLSSKQGSTK RSGLPAQQTP AQKSWIERAF YKRECVHIIP STKDPHRCC CGRLIGQHVG LTPSISVLQN EKNESRLSRN DIQSEKWSIS KHTQLSPTDA FGTIEFQGGG HSNKAMYVRV S FDTKPDLL ...String: GKKWRDAGEL ERGCSDREDS AESRRRSRSA SRGRFAESWK RLSSKQGSTK RSGLPAQQTP AQKSWIERAF YKRECVHIIP STKDPHRCC CGRLIGQHVG LTPSISVLQN EKNESRLSRN DIQSEKWSIS KHTQLSPTDA FGTIEFQGGG HSNKAMYVRV S FDTKPDLL LHLMTKEWQL ELPKLLISVH GGLQNFELQP KLKQVFGKGL IKAAMTTGAW IFTGGVNTGV IRHVGDALKD HA SKSRGKI CTIGIAPWGI VENQEDLIGR DVVRPYQTMS NPMSKLTVLN SMHSHFILAD NGTTGKYGAE VKLRRQLEKH ISL QKINTR IGQGVPVVAL IVEGGPNVIS IVLEYLRDTP PVPVVVCDGS GRASDILAFG HKYSEEGGLI NESLRDQLLV TIQK TFTYT RTQAQHLFII LMECMKKKEL ITVFRMGSEG HQDIDLAILT ALLKGANASA PDQLSLALAW NRVDIARSQI FIYGQ QWPV GSLEQAMLDA LVLDRVDFVK LLIENGVSMH RFLTISRLEE LYNTRHGPSN TLYHLVRDVK KGNLPPDYRI SLIDIG LVI EYLMGGAYRC NYTRKRFRTL YHNLFGPKRP KALKLLGMED DIPLRRGRKT TKKREEEVDI DLDDPEINHF PFPFHEL MV WAVLMKRQKM ALFFWQHGEE AMAKALVACK LCKAMAHEAS ENDMVDDISQ ELNHNSRDFG QLAVELLDQS YKQDEQLA M KLLTYELKNW SNATCLQLAV AAKHRDFIAH TCSQMLLTDM WMGRLRMRKN SGLKVILGIL LPPSILSLEF KNKDDMPYM TQAQEIHLQE KEPEEPEKPT KEKDEEDMEL TAMLGRSNGE SSRKKDEEEV QSRHRLIPVG RKIYEFYNAP IVKFWFYTLA YIGYLMLFN YIVLVKMERW PSTQEWIVIS YIFTLGIEKM REILMSEPGK LLQKVKVWLQ EYWNVTDLIA ILLFSVGMIL R LQDQPFRS DGRVIYCVNI IYWYIRLLDI FGVNKYLGPY VMMIGKMMID MMYFVIIMLV VLMSFGVARQ AILFPNEEPS WK LAKNIFY MPYWMIYGEV FADQIDPPCG QNETREDGKT IQLPPCKTGA WIVPAIMACY LLVANILLVN LLIAVFNNTF FEV KSISNQ VWKFQRYQLI MTFHERPVLP PPLIIFSHMT MIFQHVCCRW RKHESDQDER DYGLKLFITD DELKKVHDFE EQCI EEYFR EKDDRFNSSN DERIRVTSER VENMSMRLEE VNEREHSMKA SLQTVDIRLA QLEDLIGRMA TALERLTGLE RAESN KIRS RTSSDCTDAA YIVRQSSFNS QEGNTFKLQE SIDPAGEETI SPTSPTLMPR MRSHSFYSV UniProtKB: Transient receptor potential cation channel subfamily M member 3 |
-Macromolecule #2: Unidentified segment at the N-terminus of TRPM3
| Macromolecule | Name: Unidentified segment at the N-terminus of TRPM3 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.464797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
-Macromolecule #3: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE
| Macromolecule | Name: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: 3PH |
|---|---|
| Molecular weight | Theoretical: 704.998 Da |
| Chemical component information |  ChemComp-3PH: |
-Macromolecule #4: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]sp...
| Macromolecule | Name: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en type: ligand / ID: 4 / Number of copies: 4 / Formula: 9Z9 |
|---|---|
| Molecular weight | Theoretical: 544.805 Da |
| Chemical component information |  ChemComp-9Z9: |
-Macromolecule #5: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(o...
| Macromolecule | Name: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate type: ligand / ID: 5 / Number of copies: 4 / Formula: PIO |
|---|---|
| Molecular weight | Theoretical: 746.566 Da |
| Chemical component information |  ChemComp-PIO: |
-Macromolecule #6: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 6 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 53.865 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)