+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
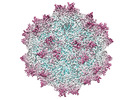
| Title | Adeno-associated virus type 2 VLP displaying M2 peptide | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mimotopes / Dengue / Zika / VIRUS LIKE PARTICLE | |||||||||
| Biological species |  adeno-associated virus 2 adeno-associated virus 2 | |||||||||
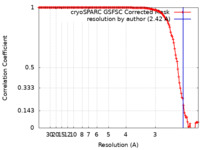
| Method | single particle reconstruction / cryo EM / Resolution: 2.42 Å | |||||||||
 Authors Authors | Hernandez C / Kopylov M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biotechnol Bioeng / Year: 2023 Journal: Biotechnol Bioeng / Year: 2023Title: Mimotope discovery as a tool to design a vaccine against Zika and dengue viruses. Authors: Esmeralda Cuevas-Juárez / Arturo Liñan-Torres / Carolina Hernández / Mykhailo Kopylov / Clint S Potter / Bridget Carragher / Octavio T Ramírez / Laura A Palomares /   Abstract: Vaccine development against dengue virus is challenging because of the antibody-dependent enhancement of infection (ADE), which causes severe disease. Consecutive infections by Zika (ZIKV) and/or ...Vaccine development against dengue virus is challenging because of the antibody-dependent enhancement of infection (ADE), which causes severe disease. Consecutive infections by Zika (ZIKV) and/or dengue viruses (DENV), or vaccination can predispose to ADE. Current vaccines and vaccine candidates contain the complete envelope viral protein, with epitopes that can raise antibodies causing ADE. We used the envelope dimer epitope (EDE), which induces neutralizing antibodies that do not elicit ADE, to design a vaccine against both flaviviruses. However, EDE is a discontinuous quaternary epitope that cannot be isolated from the E protein without other epitopes. Utilizing phage display, we selected three peptides that mimic the EDE. Free mimotopes were disordered and did not elicit an immune response. After their display on adeno-associated virus (AAV) capsids (VLP), they recovered their structure and were recognized by an EDE-specific antibody. Characterization by cryo-EM and enzyme-linked immunosorbent assay confirmed the correct display of a mimotope on the surface of the AAV VLP and its recognition by the specific antibody. Immunization with the AAV VLP displaying one of the mimotopes induced antibodies that recognized ZIKV and DENV. This work provides the basis for developing a Zika and dengue virus vaccine candidate that will not induce ADE. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28022.map.gz emd_28022.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28022-v30.xml emd-28022-v30.xml emd-28022.xml emd-28022.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28022_fsc.xml emd_28022_fsc.xml | 13.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_28022.png emd_28022.png | 151.3 KB | ||
| Masks |  emd_28022_msk_1.map emd_28022_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Others |  emd_28022_additional_1.map.gz emd_28022_additional_1.map.gz emd_28022_half_map_1.map.gz emd_28022_half_map_1.map.gz emd_28022_half_map_2.map.gz emd_28022_half_map_2.map.gz | 230.5 MB 225.1 MB 225.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28022 http://ftp.pdbj.org/pub/emdb/structures/EMD-28022 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28022 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28022 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28022.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28022.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28022_msk_1.map emd_28022_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: sharp map
| File | emd_28022_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharp map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_28022_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_28022_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : adeno-associated virus 2
| Entire | Name:  adeno-associated virus 2 adeno-associated virus 2 |
|---|---|
| Components |
|
-Supramolecule #1: adeno-associated virus 2
| Supramolecule | Name: adeno-associated virus 2 / type: virus / ID: 1 / Parent: 0 Details: Produced using the Baculovirus expression vector system and the particles were purified from the cellular pellet using an iodixanol gradient NCBI-ID: 10804 / Sci species name: adeno-associated virus 2 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: Yes |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 1935 / Average exposure time: 2.0 sec. / Average electron dose: 47.77 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)