+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | calcium channel | |||||||||
 Map data Map data | calcium channel | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cav1.3 / Channels / Calcium Ion-Selective / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvoltage-gated calcium channel activity involved SA node cell action potential / positive regulation of high voltage-gated calcium channel activity / voltage-gated calcium channel activity involved in cardiac muscle cell action potential / positive regulation of adenylate cyclase activity / Presynaptic depolarization and calcium channel opening / regulation of membrane repolarization during action potential / membrane depolarization during SA node cell action potential / calcium ion transmembrane transport via high voltage-gated calcium channel / regulation of atrial cardiac muscle cell membrane repolarization / high voltage-gated calcium channel activity ...voltage-gated calcium channel activity involved SA node cell action potential / positive regulation of high voltage-gated calcium channel activity / voltage-gated calcium channel activity involved in cardiac muscle cell action potential / positive regulation of adenylate cyclase activity / Presynaptic depolarization and calcium channel opening / regulation of membrane repolarization during action potential / membrane depolarization during SA node cell action potential / calcium ion transmembrane transport via high voltage-gated calcium channel / regulation of atrial cardiac muscle cell membrane repolarization / high voltage-gated calcium channel activity / membrane depolarization during bundle of His cell action potential / L-type voltage-gated calcium channel complex / membrane depolarization during cardiac muscle cell action potential / NCAM1 interactions / regulation of ventricular cardiac muscle cell membrane repolarization / positive regulation of calcium ion transport / cardiac muscle cell action potential involved in contraction / calcium ion import / regulation of potassium ion transmembrane transport / calcium ion transport into cytosol / Sensory processing of sound by inner hair cells of the cochlea / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / ankyrin binding / voltage-gated calcium channel complex / Mechanical load activates signaling by PIEZO1 and integrins in osteocytes / alpha-actinin binding / regulation of heart rate by cardiac conduction / calcium ion import across plasma membrane / regulation of calcium ion transport / neuronal dense core vesicle / voltage-gated calcium channel activity / presynaptic active zone membrane / sarcoplasmic reticulum / protein localization to plasma membrane / calcium channel regulator activity / Regulation of insulin secretion / sensory perception of sound / calcium ion transmembrane transport / GABA-ergic synapse / calcium channel activity / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / Z disc / cellular response to amyloid-beta / Adrenaline,noradrenaline inhibits insulin secretion / calcium ion transport / T cell receptor signaling pathway / chemical synaptic transmission / synapse / extracellular exosome / metal ion binding / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Gao S / Yao X / Yan N | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca channels. Authors: Xia Yao / Shuai Gao / Jixin Wang / Zhangqiang Li / Jian Huang / Yan Wang / Zhifei Wang / Jiaofeng Chen / Xiao Fan / Weipeng Wang / Xueqin Jin / Xiaojing Pan / Yong Yu / Armando Lagrutta / Nieng Yan /   Abstract: Drug-drug interaction of the antiviral sofosbuvir and the antiarrhythmics amiodarone has been reported to cause fatal heartbeat slowing. Sofosbuvir and its analog, MNI-1, were reported to potentiate ...Drug-drug interaction of the antiviral sofosbuvir and the antiarrhythmics amiodarone has been reported to cause fatal heartbeat slowing. Sofosbuvir and its analog, MNI-1, were reported to potentiate the inhibition of cardiomyocyte calcium handling by amiodarone, which functions as a multi-channel antagonist, and implicate its inhibitory effect on L-type Ca channels, but the molecular mechanism has remained unclear. Here we present systematic cryo-EM structural analysis of Ca1.1 and Ca1.3 treated with amiodarone or sofosbuvir alone, or sofosbuvir/MNI-1 combined with amiodarone. Whereas amiodarone alone occupies the dihydropyridine binding site, sofosbuvir is not found in the channel when applied on its own. In the presence of amiodarone, sofosbuvir/MNI-1 is anchored in the central cavity of the pore domain through specific interaction with amiodarone and directly obstructs the ion permeation path. Our study reveals the molecular basis for the physical, pharmacodynamic interaction of two drugs on the scaffold of Ca channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27907.map.gz emd_27907.map.gz | 78.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27907-v30.xml emd-27907-v30.xml emd-27907.xml emd-27907.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27907.png emd_27907.png | 138.2 KB | ||
| Filedesc metadata |  emd-27907.cif.gz emd-27907.cif.gz | 8.9 KB | ||
| Others |  emd_27907_half_map_1.map.gz emd_27907_half_map_1.map.gz emd_27907_half_map_2.map.gz emd_27907_half_map_2.map.gz | 65.4 MB 65.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27907 http://ftp.pdbj.org/pub/emdb/structures/EMD-27907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27907 | HTTPS FTP |
-Validation report
| Summary document |  emd_27907_validation.pdf.gz emd_27907_validation.pdf.gz | 985.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27907_full_validation.pdf.gz emd_27907_full_validation.pdf.gz | 985.1 KB | Display | |
| Data in XML |  emd_27907_validation.xml.gz emd_27907_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_27907_validation.cif.gz emd_27907_validation.cif.gz | 15.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27907 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27907 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27907 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27907 | HTTPS FTP |
-Related structure data
| Related structure data |  8e59MC  8e56C  8e57C  8e58C  8e5aC  8e5bC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27907.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27907.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | calcium channel | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.114 Å | ||||||||||||||||||||||||||||||||||||
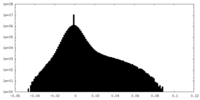
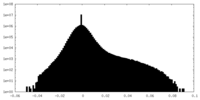
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_27907_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
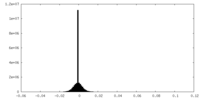
| Density Histograms |
-Half map: Half Map 2
| File | emd_27907_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
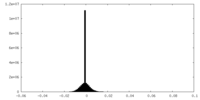
| Density Histograms |
- Sample components
Sample components
-Entire : Cav1.3
| Entire | Name: Cav1.3 |
|---|---|
| Components |
|
-Supramolecule #1: Cav1.3
| Supramolecule | Name: Cav1.3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Voltage-dependent L-type calcium channel subunit alpha-1D
| Macromolecule | Name: Voltage-dependent L-type calcium channel subunit alpha-1D type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 245.417734 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MMMMMMMKKM QHQRQQQADH ANEANYARGT RLPLSGEGPT SQPNSSKQTV LSWQAAIDAA RQAKAAQTMS TSAPPPVGSL SQRKRQQYA KSKKQGNSSN SRPARALFCL SLNNPIRRAC ISIVEWKPFD IFILLAIFAN CVALAIYIPF PEDDSNSTNH N LEKVEYAF ...String: MMMMMMMKKM QHQRQQQADH ANEANYARGT RLPLSGEGPT SQPNSSKQTV LSWQAAIDAA RQAKAAQTMS TSAPPPVGSL SQRKRQQYA KSKKQGNSSN SRPARALFCL SLNNPIRRAC ISIVEWKPFD IFILLAIFAN CVALAIYIPF PEDDSNSTNH N LEKVEYAF LIIFTVETFL KIIAYGLLLH PNAYVRNGWN LLDFVIVIVG LFSVILEQLT KETEGGNHSS GKSGGFDVKA LR AFRVLRP LRLVSGVPSL QVVLNSIIKA MVPLLHIALL VLFVIIIYAI IGLELFIGKM HKTCFFADSD IVAEEDPAPC AFS GNGRQC TANGTECRSG WVGPNGGITN FDNFAFAMLT VFQCITMEGW TDVLYWMNDA MGFELPWVYF VSLVIFGSFF VLNL VLGVL SGEFSKEREK AKARGDFQKL REKQQLEEDL KGYLDWITQA EDIDPENEEE GGEEGKRNTS MPTSETESVN TENVS GEGE NRGCCGSLCQ AISKSKLSRR WRRWNRFNRR RCRAAVKSVT FYWLVIVLVF LNTLTISSEH YNQPDWLTQI QDIANK VLL ALFTCEMLVK MYSLGLQAYF VSLFNRFDCF VVCGGITETI LVELEIMSPL GISVFRCVRL LRIFKVTRHW TSLSNLV AS LLNSMKSIAS LLLLLFLFII IFSLLGMQLF GGKFNFDETQ TKRSTFDNFP QALLTVFQIL TGEDWNAVMY DGIMAYGG P SSSGMIVCIY FIILFICGNY ILLNVFLAIA VDNLADAESL NTAQKEEAEE KERKKIARKE SLENKKNNKP EVNQIANSD NKVTIDDYRE EDEDKDPYPP CDVPVGEEEE EEEEDEPEVP AGPRPRRISE LNMKEKIAPI PEGSAFFILS KTNPIRVGCH KLINHHIFT NLILVFIMLS SAALAAEDPI RSHSFRNTIL GYFDYAFTAI FTVEILLKMT TFGAFLHKGA FCRNYFNLLD M LVVGVSLV SFGIQSSAIS VVKILRVLRV LRPLRAINRA KGLKHVVQCV FVAIRTIGNI MIVTTLLQFM FACIGVQLFK GK FYRCTDE AKSNPEECRG LFILYKDGDV DSPVVRERIW QNSDFNFDNV LSAMMALFTV STFEGWPALL YKAIDSNGEN IGP IYNHRV EISIFFIIYI IIVAFFMMNI FVGFVIVTFQ EQGEKEYKNC ELDKNQRQCV EYALKARPLR RYIPKNPYQY KFWY VVNSS PFEYMMFVLI MLNTLCLAMQ HYEQSKMFND AMDILNMVFT GVFTVEMVLK VIAFKPKGYF SDAWNTFDSL IVIGS IIDV ALSEADPTES ENVPVPTATP GNSEESNRIS ITFFRLFRVM RLVKLLSRGE GIRTLLWTFI KSFQALPYVA LLIAML FFI YAVIGMQMFG KVAMRDNNQI NRNNNFQTFP QAVLLLFRCA TGEAWQEIML ACLPGKLCDP ESDYNPGEEY TCGSNFA IV YFISFYMLCA FLIINLFVAV IMDNFDYLTR DWSILGPHHL DEFKRIWSEY DPEAKGRIKH LDVVTLLRRI QPPLGFGK L CPHRVACKRL VAMNMPLNSD GTVMFNATLF ALVRTALKIK TEGNLEQANE ELRAVIKKIW KKTSMKLLDQ VVPPAGDDE VTVGKFYATF LIQDYFRKFK KRKEQGLVGK YPAKNTTIAL QAGLRTLHDI GPEIRRAISC DLQDDEPEET KREEEDDVFK RNGALLGNH VNHVNSDRRD SLQQTNTTHR PLHVQRPSIP PASDTEKPLF PPAGNSVCHN HHNHNSIGKQ VPTSTNANLN N ANMSKAAH GKRPSIGNLE HVSENGHHSS HKHDREPQRR SSVKRTRYYE TYIRSDSGDE QLPTICREDP EIHGYFRDPH CL GEQEYFS SEECYEDDSS PTWSRQNYGY YSRYPGRNID SERPRGYHHP QGFLEDDDSP VCYDSRRSPR RRLLPPTPAS HRR SSFNFE CLRRQSSQEE VPSSPIFPHR TALPLHLMQQ QIMAVAGLDS SKAQKYSPSH STRSWATPPA TPPYRDWTPC YTPL IQVEQ SEALDQVNGS LPSLHRSSWY TDEPDISYRT FTPASLTVPS SFRNKNSDKQ RSADSLVEAV LISEGLGRYA RDPKF VSAT KHEIADACDL TIDEMESAAS TLLNGNVRPR ANGDVGPLSH RQDYELQDFG PGYSDEEPDP GRDEEDLADE MICITT L UniProtKB: Voltage-dependent L-type calcium channel subunit alpha-1D |
-Macromolecule #2: Voltage-dependent calcium channel subunit alpha-2/delta-1
| Macromolecule | Name: Voltage-dependent calcium channel subunit alpha-2/delta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 124.692469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAGCLLALT LTLFQSLLIG PSSEEPFPSA VTIKSWVDKM QEDLVTLAKT ASGVNQLVDI YEKYQDLYTV EPNNARQLVE IAARDIEKL LSNRSKALVR LALEAEKVQA AHQWREDFAS NEVVYYNAKD DLDPEKNDSE PGSQRIKPVF IEDANFGRQI S YQHAAVHI ...String: MAAGCLLALT LTLFQSLLIG PSSEEPFPSA VTIKSWVDKM QEDLVTLAKT ASGVNQLVDI YEKYQDLYTV EPNNARQLVE IAARDIEKL LSNRSKALVR LALEAEKVQA AHQWREDFAS NEVVYYNAKD DLDPEKNDSE PGSQRIKPVF IEDANFGRQI S YQHAAVHI PTDIYEGSTI VLNELNWTSA LDEVFKKNRE EDPSLLWQVF GSATGLARYY PASPWVDNSR TPNKIDLYDV RR RPWYIQG AASPKDMLIL VDVSGSVSGL TLKLIRTSVS EMLETLSDDD FVNVASFNSN AQDVSCFQHL VQANVRNKKV LKD AVNNIT AKGITDYKKG FSFAFEQLLN YNVSRANCNK IIMLFTDGGE ERAQEIFNKY NKDKKVRVFT FSVGQHNYDR GPIQ WMACE NKGYYYEIPS IGAIRINTQE YLDVLGRPMV LAGDKAKQVQ WTNVYLDALE LGLVITGTLP VFNITGQFEN KTNLK NQLI LGVMGVDVSL EDIKRLTPRF TLCPNGYYFA IDPNGYVLLH PNLQPKPIGV GIPTINLRKR RPNIQNPKSQ EPVTLD FLD AELENDIKVE IRNKMIDGES GEKTFRTLVK SQDERYIDKG NRTYTWTPVN GTDYSLALVL PTYSFYYIKA KLEETIT QA RYSETLKPDN FEESGYTFIA PRDYCNDLKI SDNNTEFLLN FNEFIDRKTP NNPSCNADLI NRVLLDAGFT NELVQNYW S KQKNIKGVKA RFVVTDGGIT RVYPKEAGEN WQENPETYED SFYKRSLDND NYVFTAPYFN KSGPGAYESG IMVSKAVEI YIQGKLLKPA VVGIKIDVNS WIENFTKTSI RDPCAGPVCD CKRNSDVMDC VILDDGGFLL MANHDDYTNQ IGRFFGEIDP SLMRHLVNI SVYAFNKSYD YQSVCEPGAA PKQGAGHRSA YVPSVADILQ IGWWATAAAW SILQQFLLSL TFPRLLEAVE M EDDDFTAS LSKQSCITEQ TQYFFDNDSK SFSGVLDCGN CSRIFHGEKL MNTNLIFIMV ESKGTCPCDT RLLIQAEQTS DG PNPCDMV KQPRYRKGPD VCFDNNVLED YTDCGGVSGL NPSLWYIIGI QFLLLWLVSG STHRLL UniProtKB: Voltage-dependent calcium channel subunit alpha-2/delta-1 |
-Macromolecule #3: Voltage-dependent L-type calcium channel subunit beta-3
| Macromolecule | Name: Voltage-dependent L-type calcium channel subunit beta-3 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.607852 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYDDSYVPGF EDSEAGSADS YTSRPSLDSD VSLEEDRESA RREVESQAQQ QLERAKHKPV AFAVRTNVSY CGVLDEECPV QGSGVNFEA KDFLHIKEKY SNDWWIGRLV KEGGDIAFIP SPQRLESIRL KQEQKARRSG NPSSLSDIGN RRSPPPSLAK Q KQKQAEHV ...String: MYDDSYVPGF EDSEAGSADS YTSRPSLDSD VSLEEDRESA RREVESQAQQ QLERAKHKPV AFAVRTNVSY CGVLDEECPV QGSGVNFEA KDFLHIKEKY SNDWWIGRLV KEGGDIAFIP SPQRLESIRL KQEQKARRSG NPSSLSDIGN RRSPPPSLAK Q KQKQAEHV PPYDVVPSMR PVVLVGPSLK GYEVTDMMQK ALFDFLKHRF DGRISITRVT ADLSLAKRSV LNNPGKRTII ER SSARSSI AEVQSEIERI FELAKSLQLV VLDADTINHP AQLAKTSLAP IIVFVKVSSP KVLQRLIRSR GKSQMKHLTV QMM AYDKLV QCPPESFDVI LDENQLEDAC EHLAEYLEVY WRATHHPAPG PGLLGPPSAI PGLQNQQLLG ERGEEHSPLE RDSL MPSDE ASESSRQAWT GSSQRSSRHL EEDYADAYQD LYQPHRQHTS GLPSANGHDP QDRLLAQDSE HNHSDRNWQR NRPWP KDSY UniProtKB: Voltage-dependent L-type calcium channel subunit beta-3 |
-Macromolecule #7: (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodo...
| Macromolecule | Name: (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone type: ligand / ID: 7 / Number of copies: 1 / Formula: BBI |
|---|---|
| Molecular weight | Theoretical: 645.312 Da |
| Chemical component information | 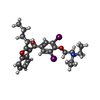 ChemComp-BBI: |
-Macromolecule #8: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 8 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #9: 1,2-Distearoyl-sn-glycerophosphoethanolamine
| Macromolecule | Name: 1,2-Distearoyl-sn-glycerophosphoethanolamine / type: ligand / ID: 9 / Number of copies: 1 / Formula: 3PE |
|---|---|
| Molecular weight | Theoretical: 748.065 Da |
| Chemical component information |  ChemComp-3PE: |
-Macromolecule #10: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 10 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.9000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)