[English] 日本語
 Yorodumi
Yorodumi- EMDB-27899: Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA | |||||||||
 Map data Map data | Half map 1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | acyltransferase / LPA / monotopic / mitochondrial / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationglycerol-3-phosphate 1-O-acyltransferase / glycerol-3-phosphate O-acyltransferase activity / phosphatidylglycerol biosynthetic process / CDP-diacylglycerol biosynthetic process / Triglyceride biosynthesis / negative regulation of activation-induced cell death of T cells / diacylglycerol biosynthetic process / triglyceride biosynthetic process / glycerophospholipid metabolic process / phosphatidic acid biosynthetic process ...glycerol-3-phosphate 1-O-acyltransferase / glycerol-3-phosphate O-acyltransferase activity / phosphatidylglycerol biosynthetic process / CDP-diacylglycerol biosynthetic process / Triglyceride biosynthesis / negative regulation of activation-induced cell death of T cells / diacylglycerol biosynthetic process / triglyceride biosynthetic process / glycerophospholipid metabolic process / phosphatidic acid biosynthetic process / Synthesis of PA / activation-induced cell death of T cells / acyl-CoA metabolic process / phospholipid homeostasis / positive regulation of multicellular organism growth / activated T cell proliferation / RUNX1 regulates estrogen receptor mediated transcription / positive regulation of activated T cell proliferation / fatty acid homeostasis / response to glucose / regulation of cytokine production / Activation of gene expression by SREBF (SREBP) / fatty acid metabolic process / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / defense response to virus / Estrogen-dependent gene expression / mitochondrial outer membrane / mitochondrion / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.67 Å | |||||||||
 Authors Authors | Wasilko DJ / Johnson ZL / Ammirati M / Chang JS / Han S / Wu H | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2023 Journal: Nat.Struct.Mol.Biol. / Year: 2023Title: Structural basis of the acyl-transfer mechanism of human GPAT1. Authors: Johnson ZL / Ammirati M / Wasilko DJ / Chang JS / Noell S / Foley TL / Yoon H / Smith K / Asano S / Hales K / Wan M / Yang Q / Piotrowski MA / Farley KA / Gilbert T / Aschenbrenner LM / ...Authors: Johnson ZL / Ammirati M / Wasilko DJ / Chang JS / Noell S / Foley TL / Yoon H / Smith K / Asano S / Hales K / Wan M / Yang Q / Piotrowski MA / Farley KA / Gilbert T / Aschenbrenner LM / Fennell KF / Dutra JK / Xu M / Guo C / Varghese AE / Bellenger J / Quinn A / Am Ende CW / West GM / Griffor MC / Bennett D / Calabrese M / Steppan CM / Han S / Wu H | |||||||||
| History |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27899.map.gz emd_27899.map.gz | 202.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27899-v30.xml emd-27899-v30.xml emd-27899.xml emd-27899.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27899_fsc.xml emd_27899_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27899.png emd_27899.png | 137.2 KB | ||
| Filedesc metadata |  emd-27899.cif.gz emd-27899.cif.gz | 7.1 KB | ||
| Others |  emd_27899_half_map_1.map.gz emd_27899_half_map_1.map.gz emd_27899_half_map_2.map.gz emd_27899_half_map_2.map.gz | 171.6 MB 171 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27899 http://ftp.pdbj.org/pub/emdb/structures/EMD-27899 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27899 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27899 | HTTPS FTP |
-Validation report
| Summary document |  emd_27899_validation.pdf.gz emd_27899_validation.pdf.gz | 984.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27899_full_validation.pdf.gz emd_27899_full_validation.pdf.gz | 984.2 KB | Display | |
| Data in XML |  emd_27899_validation.xml.gz emd_27899_validation.xml.gz | 21 KB | Display | |
| Data in CIF |  emd_27899_validation.cif.gz emd_27899_validation.cif.gz | 27.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27899 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27899 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27899 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27899 | HTTPS FTP |
-Related structure data
| Related structure data | 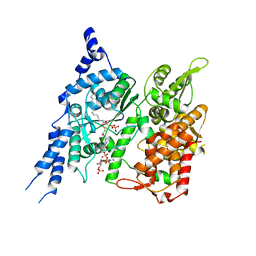 8e50MC 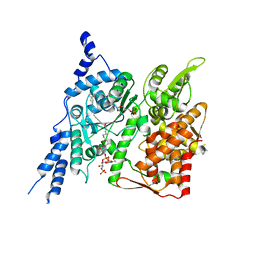 8e4yC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27899.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27899.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : glycerol-3-phosphate acyltransferase 1
| Entire | Name: glycerol-3-phosphate acyltransferase 1 |
|---|---|
| Components |
|
-Supramolecule #1: glycerol-3-phosphate acyltransferase 1
| Supramolecule | Name: glycerol-3-phosphate acyltransferase 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 86 KDa |
-Macromolecule #1: Glycerol-3-phosphate acyltransferase 1, mitochondrial
| Macromolecule | Name: Glycerol-3-phosphate acyltransferase 1, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: glycerol-3-phosphate 1-O-acyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 86.919469 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDDDDKG SENLYFQSNP SIPSLGLRNV IYINETHTRH RGWLARRLSY VLFIQERDVH KGMFATNVTE NVLNSSRVQE AIAEVAAEL NPDGSAQQQS KAVNKVKKKA KRILQEMVAT VSPAMIRLTG WVLLKLFNSF FWNIQIHKGQ LEMVKAATET N LPLLFLPV ...String: MDYKDDDDKG SENLYFQSNP SIPSLGLRNV IYINETHTRH RGWLARRLSY VLFIQERDVH KGMFATNVTE NVLNSSRVQE AIAEVAAEL NPDGSAQQQS KAVNKVKKKA KRILQEMVAT VSPAMIRLTG WVLLKLFNSF FWNIQIHKGQ LEMVKAATET N LPLLFLPV HRSHIDYLLL TFILFCHNIK APYIASGNNL NIPIFSTLIH KLGGFFIRRR LDETPDGRKD VLYRALLHGH IV ELLRQQQ FLEIFLEGTR SRSGKTSCAR AGLLSVVVDT LSTNVIPDIL IIPVGISYDR IIEGHYNGEQ LGKPKKNESL WSV ARGVIR MLRKNYGCVR VDFAQPFSLK EYLESQSQKP VSALLSLEQA LLPAILPSRP SDAADEGRDT SINESRNATD ESLR RRLIA NLAEHILFTA SKSCAIMSTH IVACLLLYRH RQGIDLSTLV EDFFVMKEEV LARDFDLGFS GNSEDVVMHA IQLLG NCVT ITHTSRNDEF FITPSTTVPS VFELNFYSNG VLHVFIMEAI IACSLYAVLN KRGLGGPTST PPNLISQEQL VRKAAS LCY LLSNEGTISL PCQTFYQVCH ETVGKFIQYG ILTVAEHDDQ EDISPSLAEQ QWDKKLPEPL SWRSDEEDED SDFGEEQ RD CYLKVSQSKE HQQFITFLQR LLGPLLEAYS SAAIFVHNFS GPVPEPEYLQ KLHKYLITRT ERNVAVYAES ATYCLVKN A VKMFKDIGVF KETKQKRVSV LELSSTFLPQ CNRQKLLEYI LSFVVL UniProtKB: Glycerol-3-phosphate acyltransferase 1, mitochondrial |
-Macromolecule #2: COENZYME A
| Macromolecule | Name: COENZYME A / type: ligand / ID: 2 / Number of copies: 1 / Formula: COA |
|---|---|
| Molecular weight | Theoretical: 767.534 Da |
| Chemical component information |  ChemComp-COA: |
-Macromolecule #3: (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate
| Macromolecule | Name: (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate / type: ligand / ID: 3 / Number of copies: 1 / Formula: NKO |
|---|---|
| Molecular weight | Theoretical: 410.483 Da |
| Chemical component information | 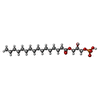 ChemComp-NKO: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.1 mg/mL | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force -5, blot time 3 sec. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 1 / Number real images: 9153 / Average exposure time: 9.0 sec. / Average electron dose: 78.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





