[English] 日本語
 Yorodumi
Yorodumi- EMDB-27890: Mouse TRPM8 structure determined in the ligand- and PI(4,5)P2-fre... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse TRPM8 structure determined in the ligand- and PI(4,5)P2-free condition, Class III, putative desensitized state | |||||||||
 Map data Map data | Full map, sharpened with B-factor -60 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRPM8 / menthol receptor / cold receptor / cold sensor / PI(4 / 5)P2 / cooling agonists / temperature sensing / ion channel / sensory transduction / transient receptor potential ion channel / MEMBRANE PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.43 Å | |||||||||
 Authors Authors | Yin Y / Zhang F / Feng S / Butay KJ / Borgnia MJ / Im W / Lee S-Y | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP. Authors: Ying Yin / Feng Zhang / Shasha Feng / Kevin John Butay / Mario J Borgnia / Wonpil Im / Seok-Yong Lee /  Abstract: The transient receptor potential melastatin 8 (TRPM8) channel is the primary molecular transducer responsible for the cool sensation elicited by menthol and cold in mammals. TRPM8 activation is ...The transient receptor potential melastatin 8 (TRPM8) channel is the primary molecular transducer responsible for the cool sensation elicited by menthol and cold in mammals. TRPM8 activation is controlled by cooling compounds together with the membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP). Our knowledge of cold sensation and the therapeutic potential of TRPM8 for neuroinflammatory diseases and pain will be enhanced by understanding the structural basis of cooling agonist- and PIP-dependent TRPM8 activation. We present cryo-electron microscopy structures of mouse TRPM8 in closed, intermediate, and open states along the ligand- and PIP-dependent gating pathway. Our results uncover two discrete agonist sites, state-dependent rearrangements in the gate positions, and a disordered-to-ordered transition of the gate-forming S6-elucidating the molecular basis of chemically induced cool sensation in mammals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27890.map.gz emd_27890.map.gz | 63.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27890-v30.xml emd-27890-v30.xml emd-27890.xml emd-27890.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27890_fsc.xml emd_27890_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_27890.png emd_27890.png | 75.7 KB | ||
| Masks |  emd_27890_msk_1.map emd_27890_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_27890_half_map_1.map.gz emd_27890_half_map_1.map.gz emd_27890_half_map_2.map.gz emd_27890_half_map_2.map.gz | 115.8 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27890 http://ftp.pdbj.org/pub/emdb/structures/EMD-27890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27890 | HTTPS FTP |
-Validation report
| Summary document |  emd_27890_validation.pdf.gz emd_27890_validation.pdf.gz | 939.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27890_full_validation.pdf.gz emd_27890_full_validation.pdf.gz | 939.5 KB | Display | |
| Data in XML |  emd_27890_validation.xml.gz emd_27890_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_27890_validation.cif.gz emd_27890_validation.cif.gz | 24.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27890 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27890 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27890 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27890 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27890.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27890.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map, sharpened with B-factor -60 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
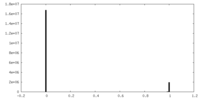
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27890_msk_1.map emd_27890_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
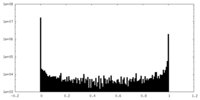
| Density Histograms |
-Half map: Half map A
| File | emd_27890_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
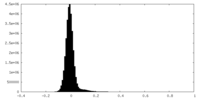
| Density Histograms |
-Half map: Half map B
| File | emd_27890_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
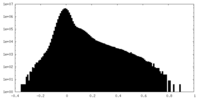
| Density Histograms |
- Sample components
Sample components
-Entire : Transient receptor potential cation channel subfamily M member 8
| Entire | Name: Transient receptor potential cation channel subfamily M member 8 |
|---|---|
| Components |
|
-Supramolecule #1: Transient receptor potential cation channel subfamily M member 8
| Supramolecule | Name: Transient receptor potential cation channel subfamily M member 8 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel subfamily M member 8
| Macromolecule | Name: Transient receptor potential cation channel subfamily M member 8 type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MASFEGARLS MRSRRNGTMG STRTLYSSVS RSTDVSYSDS DLVNFIQANF KKRECVFFTR DSKAMENICK CGYAQSQHIE GTQINQNEKW NYKKHTKEFP TDAFGDIQFE TLGKKGKYLR LSCDTDSETL YELLTQHWHL KTPNLVISVT GGAKNFALKP RMRKIFSRLI ...String: MASFEGARLS MRSRRNGTMG STRTLYSSVS RSTDVSYSDS DLVNFIQANF KKRECVFFTR DSKAMENICK CGYAQSQHIE GTQINQNEKW NYKKHTKEFP TDAFGDIQFE TLGKKGKYLR LSCDTDSETL YELLTQHWHL KTPNLVISVT GGAKNFALKP RMRKIFSRLI YIAQSKGAWI LTGGTHYGLM KYIGEVVRDN TISRNSEENI VAIGIAAWGM VSNRDTLIRS CDDEGHFSAQ YIMDDFTRDP LYILDNNHTH LLLVDNGCHG HPTVEAKLRN QLEKYISERT SQDSNYGGKI PIVCFAQGGG RETLKAINTS VKSKIPCVVV EGSGQIADVI ASLVEVEDVL TSSMVKEKLV RFLPRTVSRL PEEEIESWIK WLKEILESSH LLTVIKMEEA GDEIVSNAIS YALYKAFSTN EQDKDNWNGQ LKLLLEWNQL DLASDEIFTN DRRWESADLQ EVMFTALIKD RPKFVRLFLE NGLNLQKFLT NEVLTELFST HFSTLVYRNL QIAKNSYNDA LLTFVWKLVA NFRRSFWKED RSSREDLDVE LHDASLTTRH PLQALFIWAI LQNKKELSKV IWEQTKGCTL AALGASKLLK TLAKVKNDIN AAGESEELAN EYETRAVELF TECYSNDEDL AEQLLVYSCE AWGGSNCLEL AVEATDQHFI AQPGVQNFLS KQWYGEISRD TKNWKIILCL FIIPLVGCGL VSFRKKPIDK HKKLLWYYVA FFTSPFVVFS WNVVFYIAFL LLFAYVLLMD FHSVPHTPEL ILYALVFVLF CDEVRQWYMN GVNYFTDLWN VMDTLGLFYF IAGIVFRLHS SNKSSLYSGR VIFCLDYIIF TLRLIHIFTV SRNLGPKIIM LQRMLIDVFF FLFLFAVWMV AFGVARQGIL RQNEQRWRWI FRSVIYEPYL AMFGQVPSDV DSTTYDFSHC TFSGNESKPL CVELDEHNLP RFPEWITIPL VCIYMLSTNI LLVNLLVAMF GYTVGIVQEN NDQVWKFQRY FLVQEYCNRL NIPFPFVVFA YFYMVVKKCF KCCCKEKNME SNACCFRNED NETLAWEGVM KENYLVKINT KANDNSEEMR HRFRQLDSKL NDLKSLLKEI ANNIKSNSLE VLFQGPDYKD DDDKAHHHHH HHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 68433 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)