+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
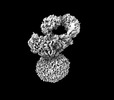
| Title | IgM BCR fab truncated form | |||||||||
 Map data Map data | IgM BCR_Delta_Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / membrane protein / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationCD22 mediated BCR regulation / riboflavin synthase activity / IgM B cell receptor complex / pentameric IgM immunoglobulin complex / B cell receptor complex / Cell surface interactions at the vascular wall / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / positive regulation of B cell activation / pre-B cell allelic exclusion / early endosome to late endosome transport ...CD22 mediated BCR regulation / riboflavin synthase activity / IgM B cell receptor complex / pentameric IgM immunoglobulin complex / B cell receptor complex / Cell surface interactions at the vascular wall / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / positive regulation of B cell activation / pre-B cell allelic exclusion / early endosome to late endosome transport / B cell affinity maturation / humoral immune response mediated by circulating immunoglobulin / riboflavin biosynthetic process / regulation of cell morphogenesis / regulation of immunoglobulin production / immunoglobulin receptor binding / immunoglobulin complex, circulating / antigen processing and presentation / immunoglobulin mediated immune response / B cell activation / B cell proliferation / positive regulation of endocytosis / antigen binding / positive regulation of B cell proliferation / multivesicular body / B cell differentiation / bioluminescence / B cell receptor signaling pathway / response to bacterium / positive regulation of immune response / transmembrane signaling receptor activity / MAPK cascade / defense response to Gram-negative bacterium / adaptive immune response / positive regulation of MAPK cascade / membrane raft / external side of plasma membrane / perinuclear region of cytoplasm / cell surface / extracellular space / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Dong Y / Pi X / Wu H / Reth M | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structural principles of B cell antigen receptor assembly. Authors: Ying Dong / Xiong Pi / Frauke Bartels-Burgahn / Deniz Saltukoglu / Zhuoyi Liang / Jianying Yang / Frederick W Alt / Michael Reth / Hao Wu /   Abstract: The B cell antigen receptor (BCR) is composed of a membrane-bound class M, D, G, E or A immunoglobulin for antigen recognition and a disulfide-linked Igα (also known as CD79A) and Igβ (also known ...The B cell antigen receptor (BCR) is composed of a membrane-bound class M, D, G, E or A immunoglobulin for antigen recognition and a disulfide-linked Igα (also known as CD79A) and Igβ (also known as CD79B) heterodimer (Igα/β) that functions as the signalling entity through intracellular immunoreceptor tyrosine-based activation motifs (ITAMs). The organizing principle of the BCR remains unknown. Here we report cryo-electron microscopy structures of mouse full-length IgM BCR and its Fab-deleted form. At the ectodomain (ECD), the Igα/β heterodimer mainly uses Igα to associate with Cµ3 and Cµ4 domains of one heavy chain (µHC) while leaving the other heavy chain (µHC') unbound. The transmembrane domain (TMD) helices of µHC and µHC' interact with those of the Igα/β heterodimer to form a tight four-helix bundle. The asymmetry at the TMD prevents the recruitment of two Igα/β heterodimers. Notably, the connecting peptide between the ECD and TMD of µHC intervenes in between those of Igα and Igβ to guide TMD assembly through charge complementarity. Weaker but distinct density for the Igβ ITAM nestles next to the TMD, suggesting potential autoinhibition of ITAM phosphorylation. Interfacial analyses suggest that all BCR classes utilize a general organizational architecture. Our studies provide a structural platform for understanding B cell signalling and designing rational therapies against BCR-mediated diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27888.map.gz emd_27888.map.gz | 129.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27888-v30.xml emd-27888-v30.xml emd-27888.xml emd-27888.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27888.png emd_27888.png | 58.9 KB | ||
| Filedesc metadata |  emd-27888.cif.gz emd-27888.cif.gz | 7.1 KB | ||
| Others |  emd_27888_half_map_1.map.gz emd_27888_half_map_1.map.gz emd_27888_half_map_2.map.gz emd_27888_half_map_2.map.gz | 127.1 MB 127.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27888 http://ftp.pdbj.org/pub/emdb/structures/EMD-27888 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27888 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27888 | HTTPS FTP |
-Related structure data
| Related structure data |  8e4cMC  8emaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27888.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27888.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | IgM BCR_Delta_Fab | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: IgM BCR Delta Fab
| File | emd_27888_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | IgM BCR_Delta_Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: IgM BCR Delta Fab
| File | emd_27888_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | IgM BCR_Delta_Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BCR complex
| Entire | Name: BCR complex |
|---|---|
| Components |
|
-Supramolecule #1: BCR complex
| Supramolecule | Name: BCR complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Isoform 2 of Immunoglobulin heavy constant mu
| Macromolecule | Name: Isoform 2 of Immunoglobulin heavy constant mu / type: protein_or_peptide / ID: 1 Details: B1-8 leader sequence: MGWSCIILFLVATATGVHS Strep Tag: WSHPQFEK Rigid linker: AEAAAKEAAAKEAAAKA Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.753785 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGWSCIILFL VATATGVHSG GWSHPQFEKA EAAAKEAAAK EAAAKAAVAE MNPNVNVFVP PRDGFSGPAP RKSKLICEAT NFTPKPITV SWLKDGKLVE SGFTTDPVTI ENKGSTPQTY KVISTLTISE IDWLNLNVYT CRVDHRGLTF LKNVSSTCAA S PSTDILTF ...String: MGWSCIILFL VATATGVHSG GWSHPQFEKA EAAAKEAAAK EAAAKAAVAE MNPNVNVFVP PRDGFSGPAP RKSKLICEAT NFTPKPITV SWLKDGKLVE SGFTTDPVTI ENKGSTPQTY KVISTLTISE IDWLNLNVYT CRVDHRGLTF LKNVSSTCAA S PSTDILTF TIPPSFADIF LSKSANLTCL VSNLATYETL NISWASQSGE PLETKIKIME SHPNGTFSAK GVASVCVEDW NN RKEFVCT VTHRDLPSPQ KKFISKPNEV HKHPPAVYLL PPAREQLNLR ESATVTCLVK GFSPADISVQ WLQRGQLLPQ EKY VTSAPM PEPGAPGFYF THSILTVTEE EWNSGETYTC VVGHEALPHL VTERTVDKST EGEVNAEEEG FENLWTTAST FIVL FLLSL FYSTTVTLFK VK UniProtKB: Immunoglobulin heavy constant mu |
-Macromolecule #2: B-cell antigen receptor complex-associated protein alpha chain,Ye...
| Macromolecule | Name: B-cell antigen receptor complex-associated protein alpha chain,Yellow fluorescent protein type: protein_or_peptide / ID: 2 Details: Flag tag:DYKDDDDK Linker: RSIATRS,Flag tag:DYKDDDDK Linker: RSIATRS YFP: ...Details: Flag tag:DYKDDDDK Linker: RSIATRS,Flag tag:DYKDDDDK Linker: RSIATRS YFP:MFKGIVEGIGIIEKIDIYTDLDKYAIRFPENMLNGIKKESSIMFNGCFLTVTSVNSNIVWFDIFEKEARKLDTFREYKVGDRVNLGTFPKFGAASGGHILSARISCVASIIEIIENEDYQQMWIQIPENFTEFLIDKDYIAVDGISLTIDTIKNNQFFISLPLKIAQNTNMKWRKKGDKVNVELSNKINANQCW,Flag Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.83723 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDKMP GGLEALRALP LLLFLSYACL GPGCQALRVE GGPPSLTVNL GEEARLTCEN NGRNPNITWW FSLQSNITWP PVPLGPGQG TTGQLFFPEV NKNHRGLYWC QVIENNILKR SCGTYLRVRN PVPRPFLDMG EGTKNRIITA EGIILLFCAV V PGTLLLFR ...String: DYKDDDDKMP GGLEALRALP LLLFLSYACL GPGCQALRVE GGPPSLTVNL GEEARLTCEN NGRNPNITWW FSLQSNITWP PVPLGPGQG TTGQLFFPEV NKNHRGLYWC QVIENNILKR SCGTYLRVRN PVPRPFLDMG EGTKNRIITA EGIILLFCAV V PGTLLLFR KRWQNEKFGR SIATRSMFKG IVEGIGIIEK IDIYTDLDKY AIRFPENMLN GIKKESSIMF NGCFLTVTSV NS NIVWFDI FEKEARKLDT FREYKVGDRV NLGTFPKFGA ASGGHILSAR ISCVASIIEI IENEDYQQMW IQIPENFTEF LID KDYIAV DGISLTIDTI KNNQFFISLP LKIAQNTNMK WRKKGDKVNV ELSNKINANQ CW UniProtKB: B-cell antigen receptor complex-associated protein alpha chain, Yellow fluorescent protein |
-Macromolecule #3: B-cell antigen receptor complex-associated protein beta chain
| Macromolecule | Name: B-cell antigen receptor complex-associated protein beta chain type: protein_or_peptide / ID: 3 / Details: full length cd79b / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.752375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATLVLSSMP CHWLLFLLLL FSGEPVPAMT SSDLPLNFQG SPCSQIWQHP RFAAKKRSSM VKFHCYTNHS GALTWFRKRG SQQPQELVS EEGRIVQTQN GSVYTLTIQN IQYEDNGIYF CKQKCDSANH NVTDSCGTEL LVLGFSTLDQ LKRRNTLKDG I ILIQTLLI ...String: MATLVLSSMP CHWLLFLLLL FSGEPVPAMT SSDLPLNFQG SPCSQIWQHP RFAAKKRSSM VKFHCYTNHS GALTWFRKRG SQQPQELVS EEGRIVQTQN GSVYTLTIQN IQYEDNGIYF CKQKCDSANH NVTDSCGTEL LVLGFSTLDQ LKRRNTLKDG I ILIQTLLI ILFIIVPIFL LLDKDDGKAG MEEDHTYEGL NIDQTATYED IVTLRTGEVK WSVGEHPGQE UniProtKB: B-cell antigen receptor complex-associated protein beta chain |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Details: full length cd79b / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)