[English] 日本語
 Yorodumi
Yorodumi- EMDB-27660: cryoEM map of de novo hallucinated homooligomer of C33-C3 symmetr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM map of de novo hallucinated homooligomer of C33-C3 symmetry, design HALC33-3_343 | ||||||||||||
 Map data Map data | EM map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | De novo protein design / Computational protein design / Machine learning / Hallucination / homo-oligomers / DE NOVO PROTEIN | ||||||||||||
| Biological species |   | ||||||||||||
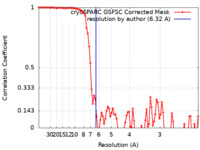
| Method | single particle reconstruction / cryo EM / Resolution: 6.32 Å | ||||||||||||
 Authors Authors | Wicky BIM / Milles LF / Courbet A / Baker D | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Hallucinating symmetric protein assemblies. Authors: B I M Wicky / L F Milles / A Courbet / R J Ragotte / J Dauparas / E Kinfu / S Tipps / R D Kibler / M Baek / F DiMaio / X Li / L Carter / A Kang / H Nguyen / A K Bera / D Baker /  Abstract: Deep learning generative approaches provide an opportunity to broadly explore protein structure space beyond the sequences and structures of natural proteins. Here, we use deep network hallucination ...Deep learning generative approaches provide an opportunity to broadly explore protein structure space beyond the sequences and structures of natural proteins. Here, we use deep network hallucination to generate a wide range of symmetric protein homo-oligomers given only a specification of the number of protomers and the protomer length. Crystal structures of seven designs are very similar to the computational models (median root mean square deviation: 0.6 angstroms), as are three cryo-electron microscopy structures of giant 10-nanometer rings with up to 1550 residues and symmetry; all differ considerably from previously solved structures. Our results highlight the rich diversity of new protein structures that can be generated using deep learning and pave the way for the design of increasingly complex components for nanomachines and biomaterials. #1:  Journal: BioRxiv / Year: 2022 Journal: BioRxiv / Year: 2022Title: Hallucinating protein assemblies Authors: Wicky BIM / Milles LF / Courbet A / Baker D | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27660.map.gz emd_27660.map.gz | 62.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27660-v30.xml emd-27660-v30.xml emd-27660.xml emd-27660.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27660_fsc.xml emd_27660_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_27660.png emd_27660.png | 15.6 KB | ||
| Masks |  emd_27660_msk_1.map emd_27660_msk_1.map | 67 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27660.cif.gz emd-27660.cif.gz | 4.8 KB | ||
| Others |  emd_27660_half_map_1.map.gz emd_27660_half_map_1.map.gz emd_27660_half_map_2.map.gz emd_27660_half_map_2.map.gz | 61.8 MB 61.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27660 http://ftp.pdbj.org/pub/emdb/structures/EMD-27660 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27660 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27660 | HTTPS FTP |
-Validation report
| Summary document |  emd_27660_validation.pdf.gz emd_27660_validation.pdf.gz | 724.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27660_full_validation.pdf.gz emd_27660_full_validation.pdf.gz | 724.5 KB | Display | |
| Data in XML |  emd_27660_validation.xml.gz emd_27660_validation.xml.gz | 16.9 KB | Display | |
| Data in CIF |  emd_27660_validation.cif.gz emd_27660_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27660 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27660 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27660 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27660 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27660.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27660.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.122 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27660_msk_1.map emd_27660_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_27660_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_27660_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : De novo hallucinated homooligomer of C33-C3 symmetry, design HALC...
| Entire | Name: De novo hallucinated homooligomer of C33-C3 symmetry, design HALC33-3_343 |
|---|---|
| Components |
|
-Supramolecule #1: De novo hallucinated homooligomer of C33-C3 symmetry, design HALC...
| Supramolecule | Name: De novo hallucinated homooligomer of C33-C3 symmetry, design HALC33-3_343 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: HALC33-3_343
| Macromolecule | Name: HALC33-3_343 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: PSNWPEVAKY FDLGKALKPI GEGLQNLKNL KHLDLSFSFS LELYPGLPSN WPEVAKYFDL GKALKPIGEG LQNLKNLKH LDLSFSFSLE LYPGLPSNWP EVAKYFDLGK ALKPIGEGLQ NLKNLKHLDL SFSFSLELYP G LPSNWPEV AKYFDLGKAL KPIGEGLQNL ...String: PSNWPEVAKY FDLGKALKPI GEGLQNLKNL KHLDLSFSFS LELYPGLPSN WPEVAKYFDL GKALKPIGEG LQNLKNLKH LDLSFSFSLE LYPGLPSNWP EVAKYFDLGK ALKPIGEGLQ NLKNLKHLDL SFSFSLELYP G LPSNWPEV AKYFDLGKAL KPIGEGLQNL KNLKHLDLSF SFSLELYPGL PSNWPEVAKY FDLGKALKPI GE GLQNLKN LKHLDLSFSF SLELYPGLPS NWPEVAKYFD LGKALKPIGE GLQNLKNLKH LDLSFSFSLE LYP GLPSNW PEVAKYFDLG KALKPIGEGL QNLKNLKHLD LSFSFSLELY PGLPSNWPEV AKYFDLGKAL KPIG EGLQN LKNLKHLDLS FSFSLELYPG LPSNWPEVAK YFDLGKALKP IGEGLQNLKN LKHLDLSFSF SLELY PGLP SNWPEVAKYF DLGKALKPIG EGLQNLKNLK HLDLSFSFSL ELYPGLPSNW PEVAKYFDLG KALKPI GEG LQNLKNLKHL DLSFSFSLEL YPGL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.4 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X