[English] 日本語
 Yorodumi
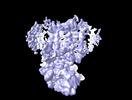
Yorodumi- EMDB-27655: Intermediate resolution structure of barley (1,3;1,4)-beta-glucan... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Intermediate resolution structure of barley (1,3;1,4)-beta-glucan synthase CslF6. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glucan / glycosyltransferase / cellulose / barley / TRANSFERASE | |||||||||
| Function / homology | plant-type cell wall organization or biogenesis / Cellulose synthase / Cellulose synthase / cellulose synthase (UDP-forming) activity / cellulose biosynthetic process / Nucleotide-diphospho-sugar transferases / cell wall organization / Golgi membrane / CslF6 Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Ho R / Purushotham P / Zimmer J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Mechanism of mixed-linkage glucan biosynthesis by barley cellulose synthase-like CslF6 (1,3;1,4)-β-glucan synthase. Authors: Pallinti Purushotham / Ruoya Ho / Long Yu / Geoffrey B Fincher / Vincent Bulone / Jochen Zimmer /    Abstract: Mixed-linkage (1,3;1,4)-β-glucans, which are widely distributed in cell walls of the grasses, are linear glucose polymers containing predominantly (1,4)-β-linked glucosyl units interspersed with ...Mixed-linkage (1,3;1,4)-β-glucans, which are widely distributed in cell walls of the grasses, are linear glucose polymers containing predominantly (1,4)-β-linked glucosyl units interspersed with single (1,3)-β-linked glucosyl units. Their distribution in cereal grains and unique structures are important determinants of dietary fibers that are beneficial to human health. We demonstrate that the barley cellulose synthase-like CslF6 enzyme is sufficient to synthesize a high-molecular weight (1,3;1,4)-β-glucan in vitro. Biochemical and cryo-electron microscopy analyses suggest that CslF6 functions as a monomer. A conserved "switch motif" at the entrance of the enzyme's transmembrane channel is critical to generate (1,3)-linkages. There, a single-point mutation markedly reduces (1,3)-linkage formation, resulting in the synthesis of cellulosic polysaccharides. Our results suggest that CslF6 monitors the orientation of the nascent polysaccharide's second or third glucosyl unit. Register-dependent interactions with these glucosyl residues reposition the polymer's terminal glucosyl unit to form either a (1,3)- or (1,4)-β-linkage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27655.map.gz emd_27655.map.gz | 92.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27655-v30.xml emd-27655-v30.xml emd-27655.xml emd-27655.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27655.png emd_27655.png | 66 KB | ||
| Filedesc metadata |  emd-27655.cif.gz emd-27655.cif.gz | 5.8 KB | ||
| Others |  emd_27655_half_map_1.map.gz emd_27655_half_map_1.map.gz emd_27655_half_map_2.map.gz emd_27655_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27655 http://ftp.pdbj.org/pub/emdb/structures/EMD-27655 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27655 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27655 | HTTPS FTP |
-Related structure data
| Related structure data |  8dqkMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27655.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27655.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
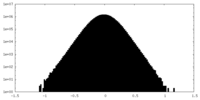
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_27655_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
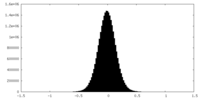
| Density Histograms |
-Half map: #1
| File | emd_27655_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
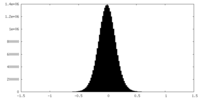
| Density Histograms |
- Sample components
Sample components
-Entire : Barley cellulose synthase-like F6
| Entire | Name: Barley cellulose synthase-like F6 |
|---|---|
| Components |
|
-Supramolecule #1: Barley cellulose synthase-like F6
| Supramolecule | Name: Barley cellulose synthase-like F6 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Cellulose synthase-like CslF6
| Macromolecule | Name: Cellulose synthase-like CslF6 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 105.193797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAPAVAGGGR VRSNEPVAAA AAAPAASGKP CVCGFQVCAC TGSAAVASAA SSLDMDIVAM GQIGAVNDES WVGVELGEDG ETDESGAAV DDRPVFRTEK IKGVLLHPYR VLIFVRLIAF TLFVIWRISH KNPDAMWLWV TSICGEFWFG FSWLLDQLPK L NPINRVPD ...String: MAPAVAGGGR VRSNEPVAAA AAAPAASGKP CVCGFQVCAC TGSAAVASAA SSLDMDIVAM GQIGAVNDES WVGVELGEDG ETDESGAAV DDRPVFRTEK IKGVLLHPYR VLIFVRLIAF TLFVIWRISH KNPDAMWLWV TSICGEFWFG FSWLLDQLPK L NPINRVPD LAVLRQRFDR PDGTSTLPGL DIFVTTADPI KEPILSTANS VLSILAADYP VDRNTCYVSD DSGMLLTYEA LA ESSKFAT LWVPFCRKHG IEPRGPESYF ELKSHPYMGR AQDEFVNDRR RVRKEYDEFK ARINSLEHDI KQRNDGYNAA IAH SQGVPR PTWMADGTQW EGTWVDASEN HRRGDHAGIV LVLLNHPSHR RQTGPPASAD NPLDLSGVDV RLPMLVYVSR EKRP GHDHQ KKAGAMNALT RASALLSNSP FILNLDCDHY INNSQALRAG ICFMVGRDSD TVAFVQFPQR FEGVDPTDLY ANHNR IFFD GTLRALDGMQ GPIYVGTGCL FRRITVYGFD PPRINVGGPC FPRLAGLFAK TKYEKPGLEM TTAKAKAAPV PAKGKH GFL PLPKKTYGKS DAFVDTIPRA SHPSPYAAAA EGIVADEATI VEAVNVTAAA FEKKTGWGKE IGWVYDTVTE DVVTGYR MH IKGWRSRYCS IYPHAFIGTA PINLTERLFQ VLRWSTGSLE IFFSKNNPLF GSTYLHPLQR VAYINITTYP FTAIFLIF Y TTVPALSFVT GHFIVQRPTT MFYVYLGIVL STLLVIAVLE VKWAGVTVFE WFRNGQFWMT ASCSAYLAAV CQVLTKVIF RRDISFKLTS KLPSGDEKKD PYADLYVVRW TPLMITPIII IFVNIIGSAV AFAKVLDGEW THWLKVAGGV FFNFWVLFHL YPFAKGILG KHGKTPVVVL VWWAFTFVIT AVLYINIPHM HTSGGKHTTV HGHHGKKLVD TGLYGWLH UniProtKB: CslF6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: AlphaFold2 predicted structure |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 80347 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller




 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)