[English] 日本語
 Yorodumi
Yorodumi- EMDB-27291: Human PRPS1-E307A engineered mutation with Phosphate, ATP, and R5... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human PRPS1-E307A engineered mutation with Phosphate, ATP, and R5P; Hexamer | |||||||||
 Map data Map data | Human PRPS1-E307A engineered mutation with Phosphate, ATP, and R5P, Hexamer Centered | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phosphoribosyl pyrophosphate synthetase / ATP/R5P substrate / Filament-breaking mutation / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology information5-Phosphoribose 1-diphosphate biosynthesis / hypoxanthine biosynthetic process / ribose phosphate diphosphokinase complex / ribonucleoside monophosphate biosynthetic process / ribose-phosphate diphosphokinase / ribose phosphate diphosphokinase activity / urate biosynthetic process / 5-phosphoribose 1-diphosphate biosynthetic process / pyrimidine nucleotide biosynthetic process / purine nucleotide biosynthetic process ...5-Phosphoribose 1-diphosphate biosynthesis / hypoxanthine biosynthetic process / ribose phosphate diphosphokinase complex / ribonucleoside monophosphate biosynthetic process / ribose-phosphate diphosphokinase / ribose phosphate diphosphokinase activity / urate biosynthetic process / 5-phosphoribose 1-diphosphate biosynthetic process / pyrimidine nucleotide biosynthetic process / purine nucleotide biosynthetic process / purine nucleobase metabolic process / kinase activity / nervous system development / magnesium ion binding / protein homodimerization activity / ATP binding / identical protein binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Hvorecny KL / Kollman JM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2023 Journal: Nat.Struct.Mol.Biol. / Year: 2023Title: Human PRPS1 filaments stabilize allosteric sites to regulate activity. Authors: Hvorecny KL / Hargett K / Quispe JD / Kollman JM | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27291.map.gz emd_27291.map.gz | 14.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27291-v30.xml emd-27291-v30.xml emd-27291.xml emd-27291.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27291.png emd_27291.png | 129.3 KB | ||
| Filedesc metadata |  emd-27291.cif.gz emd-27291.cif.gz | 5.8 KB | ||
| Others |  emd_27291_half_map_1.map.gz emd_27291_half_map_1.map.gz emd_27291_half_map_2.map.gz emd_27291_half_map_2.map.gz | 98.2 MB 98.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27291 http://ftp.pdbj.org/pub/emdb/structures/EMD-27291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27291 | HTTPS FTP |
-Validation report
| Summary document |  emd_27291_validation.pdf.gz emd_27291_validation.pdf.gz | 683 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27291_full_validation.pdf.gz emd_27291_full_validation.pdf.gz | 682.6 KB | Display | |
| Data in XML |  emd_27291_validation.xml.gz emd_27291_validation.xml.gz | 14.2 KB | Display | |
| Data in CIF |  emd_27291_validation.cif.gz emd_27291_validation.cif.gz | 16.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27291 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27291 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27291 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27291 | HTTPS FTP |
-Related structure data
| Related structure data |  8dbnMC  8dbcC  8dbdC  8dbeC  8dbfC  8dbgC  8dbhC  8dbiC  8dbjC  8dbkC  8dblC  8dbmC  8dboC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27291.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27291.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human PRPS1-E307A engineered mutation with Phosphate, ATP, and R5P, Hexamer Centered | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||||||||||||||||||
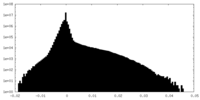
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Human PRPS1-E307A engineered mutation with Phosphate, ATP, and...
| File | emd_27291_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human PRPS1-E307A engineered mutation with Phosphate, ATP, and R5P, Hexamer Centered half 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
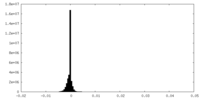
| Density Histograms |
-Half map: Human PRPS1-E307A engineered mutation with Phosphate, ATP, and...
| File | emd_27291_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human PRPS1-E307A engineered mutation with Phosphate, ATP, and R5P, Hexamer Centered half 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
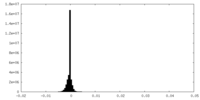
| Density Histograms |
- Sample components
Sample components
-Entire : Hexamer of phosphoribosyl pyrophosphate synthetase 1 E307A engine...
| Entire | Name: Hexamer of phosphoribosyl pyrophosphate synthetase 1 E307A engineered mutation with phosphate, ATP, and R5P |
|---|---|
| Components |
|
-Supramolecule #1: Hexamer of phosphoribosyl pyrophosphate synthetase 1 E307A engine...
| Supramolecule | Name: Hexamer of phosphoribosyl pyrophosphate synthetase 1 E307A engineered mutation with phosphate, ATP, and R5P type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Six copies of PRPS1-E307A assemble to form a hexamer. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Ribose-phosphate pyrophosphokinase 1
| Macromolecule | Name: Ribose-phosphate pyrophosphokinase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: ribose-phosphate diphosphokinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.777086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SPNIKIFSGS SHQDLSQKIA DRLGLELGKV VTKKFSNQET CVEIGESVRG EDVYIVQSGC GEINDNLMEL LIMINACKIA SASRVTAVI PCFPYARQDK KDKSRAPISA KLVANMLSVA GADHIITMDL HASQIQGFFD IPVDNLYAEP AVLKWIRENI S EWRNCTIV ...String: SPNIKIFSGS SHQDLSQKIA DRLGLELGKV VTKKFSNQET CVEIGESVRG EDVYIVQSGC GEINDNLMEL LIMINACKIA SASRVTAVI PCFPYARQDK KDKSRAPISA KLVANMLSVA GADHIITMDL HASQIQGFFD IPVDNLYAEP AVLKWIRENI S EWRNCTIV SPDAGGAKRV TSIADRLNVD FALIHKERKK ANEVDRMVLV GDVKDRVAIL VDDMADTCGT ICHAADKLLS AG ATRVYAI LTHGIFSGPA ISRINNACFE AVVVTNTIPQ EDKMKHCSKI QVIDISMILA EAIRRTHNGA SVSYLFSHVP L UniProtKB: Ribose-phosphate pyrophosphokinase 1 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 6 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: 5-O-phosphono-alpha-D-ribofuranose
| Macromolecule | Name: 5-O-phosphono-alpha-D-ribofuranose / type: ligand / ID: 3 / Number of copies: 6 / Formula: HSX |
|---|---|
| Molecular weight | Theoretical: 230.11 Da |
| Chemical component information | 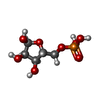 ChemComp-HSX: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 12 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: C-flat / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 90.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.75 µm / Nominal defocus min: 0.75 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D3 (2x3 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 2.4 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PHENIX (ver. 1.18) / Number images used: 810903 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8dbn: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)