+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human PRPS1 with Phosphate; Hexamer | |||||||||
 Map data Map data | Human PRPS1 with Phosphate; Hexamer Centered | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phosphoribosyl pyrophosphate synthetase / phosphate / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology information5-Phosphoribose 1-diphosphate biosynthesis / hypoxanthine biosynthetic process / ribose phosphate diphosphokinase complex / ribonucleoside monophosphate biosynthetic process / ribose-phosphate diphosphokinase / ribose phosphate diphosphokinase activity / urate biosynthetic process / 5-phosphoribose 1-diphosphate biosynthetic process / pyrimidine nucleotide biosynthetic process / purine nucleotide biosynthetic process ...5-Phosphoribose 1-diphosphate biosynthesis / hypoxanthine biosynthetic process / ribose phosphate diphosphokinase complex / ribonucleoside monophosphate biosynthetic process / ribose-phosphate diphosphokinase / ribose phosphate diphosphokinase activity / urate biosynthetic process / 5-phosphoribose 1-diphosphate biosynthetic process / pyrimidine nucleotide biosynthetic process / purine nucleotide biosynthetic process / purine nucleobase metabolic process / kinase activity / nervous system development / magnesium ion binding / protein homodimerization activity / ATP binding / identical protein binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Hvorecny KL / Kollman JM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Human PRPS1 filaments stabilize allosteric sites to regulate activity. Authors: Kelli L Hvorecny / Kenzee Hargett / Joel D Quispe / Justin M Kollman /  Abstract: The universally conserved enzyme phosphoribosyl pyrophosphate synthetase (PRPS) assembles filaments in evolutionarily diverse organisms. PRPS is a key regulator of nucleotide metabolism, and ...The universally conserved enzyme phosphoribosyl pyrophosphate synthetase (PRPS) assembles filaments in evolutionarily diverse organisms. PRPS is a key regulator of nucleotide metabolism, and mutations in the human enzyme PRPS1 lead to a spectrum of diseases. Here we determine structures of human PRPS1 filaments in active and inhibited states, with fixed assembly contacts accommodating both conformations. The conserved assembly interface stabilizes the binding site for the essential activator phosphate, increasing activity in the filament. Some disease mutations alter assembly, supporting the link between filament stability and activity. Structures of active PRPS1 filaments turning over substrate also reveal coupling of catalysis in one active site with product release in an adjacent site. PRPS1 filaments therefore provide an additional layer of allosteric control, conserved throughout evolution, with likely impact on metabolic homeostasis. Stabilization of allosteric binding sites by polymerization adds to the growing diversity of assembly-based enzyme regulatory mechanisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27279.map.gz emd_27279.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27279-v30.xml emd-27279-v30.xml emd-27279.xml emd-27279.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27279.png emd_27279.png | 109 KB | ||
| Filedesc metadata |  emd-27279.cif.gz emd-27279.cif.gz | 5.5 KB | ||
| Others |  emd_27279_half_map_1.map.gz emd_27279_half_map_1.map.gz emd_27279_half_map_2.map.gz emd_27279_half_map_2.map.gz | 67.8 MB 67.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27279 http://ftp.pdbj.org/pub/emdb/structures/EMD-27279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27279 | HTTPS FTP |
-Validation report
| Summary document |  emd_27279_validation.pdf.gz emd_27279_validation.pdf.gz | 632.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27279_full_validation.pdf.gz emd_27279_full_validation.pdf.gz | 631.9 KB | Display | |
| Data in XML |  emd_27279_validation.xml.gz emd_27279_validation.xml.gz | 13.2 KB | Display | |
| Data in CIF |  emd_27279_validation.cif.gz emd_27279_validation.cif.gz | 15.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27279 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27279 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27279 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27279 | HTTPS FTP |
-Related structure data
| Related structure data |  8dbcMC  8dbdC  8dbeC  8dbfC  8dbgC  8dbhC  8dbiC  8dbjC  8dbkC  8dblC  8dbmC  8dbnC  8dboC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27279.map.gz / Format: CCP4 / Size: 87.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27279.map.gz / Format: CCP4 / Size: 87.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human PRPS1 with Phosphate; Hexamer Centered | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.145 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Human PRPS1 with Phosphate; Hexamer Centered half 2
| File | emd_27279_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human PRPS1 with Phosphate; Hexamer Centered half 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
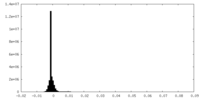
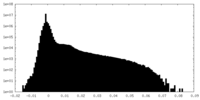
| Density Histograms |
-Half map: Human PRPS1 with Phosphate; Hexamer Centered half 1
| File | emd_27279_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human PRPS1 with Phosphate; Hexamer Centered half 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
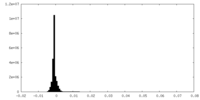
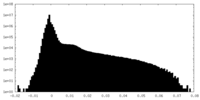
| Density Histograms |
- Sample components
Sample components
-Entire : Hexamer of phosphoribosyl pyrophosphate synthetase 1 with phosphate
| Entire | Name: Hexamer of phosphoribosyl pyrophosphate synthetase 1 with phosphate |
|---|---|
| Components |
|
-Supramolecule #1: Hexamer of phosphoribosyl pyrophosphate synthetase 1 with phosphate
| Supramolecule | Name: Hexamer of phosphoribosyl pyrophosphate synthetase 1 with phosphate type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Six copies of PRPS1 assemble to form a hexamer; hexamers assembly linearly to form filaments. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Ribose-phosphate pyrophosphokinase 1
| Macromolecule | Name: Ribose-phosphate pyrophosphokinase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: ribose-phosphate diphosphokinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.879238 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPNIKIFSGS SHQDLSQKIA DRLGLELGKV VTKKFSNQET CVEIGESVRG EDVYIVQSGC GEINDNLMEL LIMINACKIA SASRVTAVI PCFPYARQDK KDKSRAPISA KLVANMLSVA GADHIITMDL HASQIQGFFD IPVDNLYAEP AVLKWIRENI S EWRNCTIV ...String: MPNIKIFSGS SHQDLSQKIA DRLGLELGKV VTKKFSNQET CVEIGESVRG EDVYIVQSGC GEINDNLMEL LIMINACKIA SASRVTAVI PCFPYARQDK KDKSRAPISA KLVANMLSVA GADHIITMDL HASQIQGFFD IPVDNLYAEP AVLKWIRENI S EWRNCTIV SPDAGGAKRV TSIADRLNVD FALIHKERKK ANEVDRMVLV GDVKDRVAIL VDDMADTCGT ICHAADKLLS AG ATRVYAI LTHGIFSGPA ISRINNACFE AVVVTNTIPQ EDKMKHCSKI QVIDISMILA EAIRRTHNGE SVSYLFSHVP L UniProtKB: Ribose-phosphate pyrophosphokinase 1 |
-Macromolecule #2: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 2 / Number of copies: 12 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: C-flat / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.75 µm |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D3 (2x3 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PHENIX (ver. 1.18) / Number images used: 158965 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8dbc: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)