[English] 日本語
 Yorodumi
Yorodumi- EMDB-27229: Cryo-EM map of human CNTF signaling complex with full extracellul... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of human CNTF signaling complex with full extracellular domains: refined from a particle subset, with lower global resolution but improved LIFR D6-D8 density | |||||||||
 Map data Map data | CNTF_additional map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.37 Å | |||||||||
 Authors Authors | Zhou Y / Franklin MC | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structural insights into the assembly of gp130 family cytokine signaling complexes. Authors: Yi Zhou / Panayiotis E Stevis / Jing Cao / Kei Saotome / Jiaxi Wu / Arielle Glatman Zaretsky / Sokol Haxhinasto / George D Yancopoulos / Andrew J Murphy / Mark W Sleeman / William C Olson / Matthew C Franklin /  Abstract: The interleukin-6 (IL-6) family cytokines signal through gp130 receptor homodimerization or heterodimerization with a second signaling receptor and play crucial roles in various cellular processes. ...The interleukin-6 (IL-6) family cytokines signal through gp130 receptor homodimerization or heterodimerization with a second signaling receptor and play crucial roles in various cellular processes. We determined cryo-electron microscopy structures of five signaling complexes of this family, containing full receptor ectodomains bound to their respective ligands ciliary neurotrophic factor, cardiotrophin-like cytokine factor 1 (CLCF1), leukemia inhibitory factor, IL-27, and IL-6. Our structures collectively reveal similarities and differences in the assembly of these complexes. The acute bends at both signaling receptors in all complexes bring the membrane-proximal domains to a ~30 angstrom range but with distinct distances and orientations. We also reveal how CLCF1 engages its secretion chaperone cytokine receptor-like factor 1. Our data provide valuable insights for therapeutically targeting gp130-mediated signaling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27229.map.gz emd_27229.map.gz | 229 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27229-v30.xml emd-27229-v30.xml emd-27229.xml emd-27229.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27229.png emd_27229.png | 104 KB | ||
| Others |  emd_27229_half_map_1.map.gz emd_27229_half_map_1.map.gz emd_27229_half_map_2.map.gz emd_27229_half_map_2.map.gz | 226.8 MB 226.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27229 http://ftp.pdbj.org/pub/emdb/structures/EMD-27229 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27229 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27229 | HTTPS FTP |
-Validation report
| Summary document |  emd_27229_validation.pdf.gz emd_27229_validation.pdf.gz | 1015.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27229_full_validation.pdf.gz emd_27229_full_validation.pdf.gz | 1014.9 KB | Display | |
| Data in XML |  emd_27229_validation.xml.gz emd_27229_validation.xml.gz | 15.4 KB | Display | |
| Data in CIF |  emd_27229_validation.cif.gz emd_27229_validation.cif.gz | 17.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27229 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27229 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27229 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27229 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27229.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27229.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CNTF_additional map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
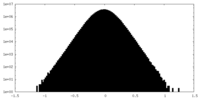
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map B
| File | emd_27229_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
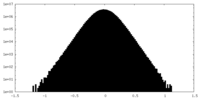
| Density Histograms |
-Half map: half map A
| File | emd_27229_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
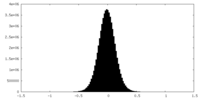
| Density Histograms |
- Sample components
Sample components
-Entire : Human CNTF in complex with CNTFR alpha, gp130, and LIFR
| Entire | Name: Human CNTF in complex with CNTFR alpha, gp130, and LIFR |
|---|---|
| Components |
|
-Supramolecule #1: Human CNTF in complex with CNTFR alpha, gp130, and LIFR
| Supramolecule | Name: Human CNTF in complex with CNTFR alpha, gp130, and LIFR type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Interleukin-6 receptor subunit beta
| Macromolecule | Name: Interleukin-6 receptor subunit beta / type: protein_or_peptide / ID: 1 Details: Models of gp130 D1 and D6 derived from PDB:1P9M and PDB:3L5H were rigid-body refined against the EM density Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 71.233203 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ELLDPCGYIS PESPVVQLHS NFTAVCVLKE KCMDYFHVNA NYIVWKTNHF TIPKEQYTII NRTASSVTFT DIASLNIQLT CNILTFGQL EQNVYGITII SGLPPEKPKN LSCIVNEGKK MRCEWDGGRE THLETNFTLK SEWATHKFAD CKAKRDTPTS C TVDYSTVY ...String: ELLDPCGYIS PESPVVQLHS NFTAVCVLKE KCMDYFHVNA NYIVWKTNHF TIPKEQYTII NRTASSVTFT DIASLNIQLT CNILTFGQL EQNVYGITII SGLPPEKPKN LSCIVNEGKK MRCEWDGGRE THLETNFTLK SEWATHKFAD CKAKRDTPTS C TVDYSTVY FVNIEVWVEA ENALGKVTSD HINFDPVYKV KPNPPHNLSV INSEELSSIL KLTWTNPSIK SVIILKYNIQ YR TKDASTW SQIPPEDTAS TRSSFTVQDL KPFTEYVFRI RCMKEDGKGY WSDWSEEASG ITYEDRPSKA PSFWYKIDPS HTQ GYRTVQ LVWKTLPPFE ANGKILDYEV TLTRWKSHLQ NYTVNATKLT VNLTNDRYLA TLTVRNLVGK SDAAVLTIPA CDFQ ATHPV MDLKAFPKDN MLWVEWTTPR ESVKKYILEW CVLSDKAPCI TDWQQEDGTV HRTYLRGNLA ESKCYLITVT PVYAD GPGS PESIKAYLKQ APPSKGPTVR TKKVGKNEAV LEWDQLPVDV QNGFIRNYTI FYRTIIGNET AVNVDSSHTE YTLSSL TSD TLYMVRMAAY TDEGGKDGPE FTFTTPKFAQ GEIEEQKLIS EEDLGGEQKL ISEEDLHHHH HH |
-Macromolecule #2: Ciliary neurotrophic factor
| Macromolecule | Name: Ciliary neurotrophic factor / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.704854 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAFTEHSPLT PHRRDLCSRS IWLARKIRSD LTALTESYVK HQGLNKNINL DSADGMPVAS TDQWSELTEA ERLQENLQAY RTFHVLLAR LLEDQQVHFT PTEGDFHQAI HTLLLQVAAF AYQIEELMIL LEYKIPRNEA DGMPINVGDG GLFEKKLWGL K VLQELSQW ...String: MAFTEHSPLT PHRRDLCSRS IWLARKIRSD LTALTESYVK HQGLNKNINL DSADGMPVAS TDQWSELTEA ERLQENLQAY RTFHVLLAR LLEDQQVHFT PTEGDFHQAI HTLLLQVAAF AYQIEELMIL LEYKIPRNEA DGMPINVGDG GLFEKKLWGL K VLQELSQW TVRSIHDLRF ISSHQTGIEQ KLISEEDLGG EQKLISEEDL HHHHHH |
-Macromolecule #3: Ciliary neurotrophic factor receptor subunit alpha
| Macromolecule | Name: Ciliary neurotrophic factor receptor subunit alpha / type: protein_or_peptide / ID: 3 Details: The model of CNTFR alpha D1 derived from PDB:8D7E was rigid-body refined against the EM density Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.855305 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QRHSPQEAPH VQYERLGSDV TLPCGTANWD AAVTWRVNGT DLAPDLLNGS QLVLHGLELG HSGLYACFHR DSWHLRHQVL LHVGLPPRE PVLSCRSNTY PKGFYCSWHL PTPTYIPNTF NVTVLHGSKI MVCEKDPALK NRCHIRYMHL FSTIKYKVSI S VSNALGHN ...String: QRHSPQEAPH VQYERLGSDV TLPCGTANWD AAVTWRVNGT DLAPDLLNGS QLVLHGLELG HSGLYACFHR DSWHLRHQVL LHVGLPPRE PVLSCRSNTY PKGFYCSWHL PTPTYIPNTF NVTVLHGSKI MVCEKDPALK NRCHIRYMHL FSTIKYKVSI S VSNALGHN ATAITFDEFT IVKPDPPENV VARPVPSNPR RLEVTWQTPS TWPDPESFPL KFFLRYRPLI LDQWQHVELS DG TAHTITD AYAGKEYIIQ VAAKDNEIGT WSDWSVAAHA TPWTEEPRHL TTEAQAAETT TSTTSSLAPP PTTKICDPGE LGS |
-Macromolecule #4: Leukemia inhibitory factor receptor
| Macromolecule | Name: Leukemia inhibitory factor receptor / type: protein_or_peptide / ID: 4 Details: Models of LIFR D6-D8 predicted by AlphaFold and LIFR D1 derived from PDB:3E0G were rigid-body refined against the EM density Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 92.834812 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QKKGAPHDLK CVTNNLQVWN CSWKAPSGTG RGTDYEVCIE NRSRSCYQLE KTSIKIPALS HGDYEITINS LHDFGSSTSK FTLNEQNVS LIPDTPEILN LSADFSTSTL YLKWNDRGSV FPHRSNVIWE IKVLRKESME LVKLVTHNTT LNGKDTLHHW S WASDMPLE ...String: QKKGAPHDLK CVTNNLQVWN CSWKAPSGTG RGTDYEVCIE NRSRSCYQLE KTSIKIPALS HGDYEITINS LHDFGSSTSK FTLNEQNVS LIPDTPEILN LSADFSTSTL YLKWNDRGSV FPHRSNVIWE IKVLRKESME LVKLVTHNTT LNGKDTLHHW S WASDMPLE CAIHFVEIRC YIDNLHFSGL EEWSDWSPVK NISWIPDSQT KVFPQDKVIL VGSDITFCCV SQEKVLSALI GH TNCPLIH LDGENVAIKI RNISVSASSG TNVVFTTEDN IFGTVIFAGY PPDTPQQLNC ETHDLKEIIC SWNPGRVTAL VGP RATSYT LVESFSGKYV RLKRAEAPTN ESYQLLFQML PNQEIYNFTL NAHNPLGRSQ STILVNITEK VYPHTPTSFK VKDI NSTAV KLSWHLPGNF AKINFLCEIE IKKSNSVQEQ RNVTIKGVEN SSYLVALDKL NPYTLYTFRI RCSTETFWKW SKWSN KKQH LTTEASPSKG PDTWREWSSD GKNLIIYWKP LPINEANGKI LSYNVSCSSD EETQSLSEIP DPQHKAEIRL DKNDYI ISV VAKNSVGSSP PSKIASMEIP NDDLKIEQVV GMGKGILLTW HYDPNMTCDY VIKWCNSSRS EPCLMDWRKV PSNSTET VI ESDEFRPGIR YNFFLYGCRN QGYQLLRSMI GYIEELAPIV APNFTVEDTS ADSILVKWED IPVEELRGFL RGYLFYFG K GERDTSKMRV LESGRSDIKV KNITDISQKT LRIADLQGKT SYHLVLRAYT DGGVGPEKSM YVVTKENSEQ KLISEEDLG GEQKLISEED LHHHHHH |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.37 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 100013 |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)