[English] 日本語
 Yorodumi
Yorodumi- EMDB-26985: Human excitatory amino acid transporter 3 (EAAT3) with bound glut... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human excitatory amino acid transporter 3 (EAAT3) with bound glutamate in an intermediate outward facing state | |||||||||
 Map data Map data | Excitatory amino acid transporter 3 with glutamate bound at new intermediate outward facing states | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationD-aspartate transmembrane transport / regulation of protein targeting to membrane / D-aspartate transmembrane transporter activity / Defective SLC1A1 is implicated in schizophrenia 18 (SCZD18) and dicarboxylic aminoaciduria (DCBXA) / distal dendrite / cysteine transport / cysteine transmembrane transporter activity / neurotransmitter receptor transport to plasma membrane / high-affinity L-glutamate transmembrane transporter activity / glutamate:sodium symporter activity ...D-aspartate transmembrane transport / regulation of protein targeting to membrane / D-aspartate transmembrane transporter activity / Defective SLC1A1 is implicated in schizophrenia 18 (SCZD18) and dicarboxylic aminoaciduria (DCBXA) / distal dendrite / cysteine transport / cysteine transmembrane transporter activity / neurotransmitter receptor transport to plasma membrane / high-affinity L-glutamate transmembrane transporter activity / glutamate:sodium symporter activity / L-glutamate import / response to decreased oxygen levels / cellular response to mercury ion / : / L-glutamate transmembrane transporter activity / retina layer formation / L-glutamate transmembrane transport / glutathione biosynthetic process / D-aspartate import across plasma membrane / L-aspartate transmembrane transport / cellular response to ammonium ion / righting reflex / zinc ion transmembrane transport / L-aspartate transmembrane transporter activity / cellular response to bisphenol A / intracellular glutamate homeostasis / grooming behavior / L-aspartate import across plasma membrane / Glutamate Neurotransmitter Release Cycle / monoatomic anion channel activity / L-glutamate import across plasma membrane / proximal dendrite / transepithelial transport / apical dendrite / intracellular zinc ion homeostasis / blood vessel morphogenesis / cellular response to cocaine / chloride transmembrane transporter activity / motor neuron apoptotic process / G protein-coupled dopamine receptor signaling pathway / response to anesthetic / response to morphine / glutamate receptor signaling pathway / superoxide metabolic process / neurotransmitter transport / heart contraction / perisynaptic space / maintenance of blood-brain barrier / dopamine metabolic process / adult behavior / motor behavior / asymmetric synapse / conditioned place preference / glial cell projection / synaptic cleft / behavioral fear response / postsynaptic modulation of chemical synaptic transmission / response to axon injury / positive regulation of heart rate / transport across blood-brain barrier / monoatomic ion transport / neurogenesis / axon terminus / chloride transmembrane transport / response to amphetamine / dendritic shaft / cell periphery / locomotory behavior / synapse organization / brain development / Schaffer collateral - CA1 synapse / memory / long-term synaptic potentiation / recycling endosome membrane / cytokine-mediated signaling pathway / late endosome membrane / presynapse / cellular response to oxidative stress / early endosome membrane / perikaryon / chemical synaptic transmission / gene expression / dendritic spine / negative regulation of neuron apoptotic process / apical plasma membrane / membrane raft / response to xenobiotic stimulus / axon / neuronal cell body / dendrite / cell surface / extracellular exosome / metal ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
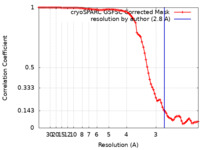
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Qiu B / Boudker O | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Symport and antiport mechanisms of human glutamate transporters. Authors: Biao Qiu / Olga Boudker /  Abstract: Excitatory amino acid transporters (EAATs) uptake glutamate into glial cells and neurons. EAATs achieve million-fold transmitter gradients by symporting it with three sodium ions and a proton, and ...Excitatory amino acid transporters (EAATs) uptake glutamate into glial cells and neurons. EAATs achieve million-fold transmitter gradients by symporting it with three sodium ions and a proton, and countertransporting a potassium ion via an elevator mechanism. Despite the availability of structures, the symport and antiport mechanisms still need to be clarified. We report high-resolution cryo-EM structures of human EAAT3 bound to the neurotransmitter glutamate with symported ions, potassium ions, sodium ions alone, or without ligands. We show that an evolutionarily conserved occluded translocation intermediate has a dramatically higher affinity for the neurotransmitter and the countertransported potassium ion than outward- or inward-facing transporters and plays a crucial role in ion coupling. We propose a comprehensive ion coupling mechanism involving a choreographed interplay between bound solutes, conformations of conserved amino acid motifs, and movements of the gating hairpin and the substrate-binding domain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26985.map.gz emd_26985.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26985-v30.xml emd-26985-v30.xml emd-26985.xml emd-26985.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26985_fsc.xml emd_26985_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_26985.png emd_26985.png | 79.1 KB | ||
| Filedesc metadata |  emd-26985.cif.gz emd-26985.cif.gz | 6.2 KB | ||
| Others |  emd_26985_half_map_1.map.gz emd_26985_half_map_1.map.gz emd_26985_half_map_2.map.gz emd_26985_half_map_2.map.gz | 59 MB 59 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26985 http://ftp.pdbj.org/pub/emdb/structures/EMD-26985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26985 | HTTPS FTP |
-Validation report
| Summary document |  emd_26985_validation.pdf.gz emd_26985_validation.pdf.gz | 750.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26985_full_validation.pdf.gz emd_26985_full_validation.pdf.gz | 750.2 KB | Display | |
| Data in XML |  emd_26985_validation.xml.gz emd_26985_validation.xml.gz | 16.6 KB | Display | |
| Data in CIF |  emd_26985_validation.cif.gz emd_26985_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26985 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26985 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26985 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26985 | HTTPS FTP |
-Related structure data
| Related structure data |  8ctcMC  8ctdC  8cuaC  8cudC  8cuiC  8cujC  8cv2C  8cv3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26985.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26985.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Excitatory amino acid transporter 3 with glutamate bound at new intermediate outward facing states | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_26985_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_26985_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human EAAT3 with glutamate bound
| Entire | Name: Human EAAT3 with glutamate bound |
|---|---|
| Components |
|
-Supramolecule #1: Human EAAT3 with glutamate bound
| Supramolecule | Name: Human EAAT3 with glutamate bound / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Human EAAT3 with glutamate bound
| Supramolecule | Name: Human EAAT3 with glutamate bound / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Excitatory amino acid transporter 3
| Macromolecule | Name: Excitatory amino acid transporter 3 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.120863 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPMGKPARKG AEWKRFLKNN WVLLSTVAAV VLGITTGVLV REHSNLSTLE KFYFAFPGEI LMRMLKLIIL PLIISSMITG VAALDSNVS GKIGLRAVVY YFATTLIAVI LGIVLVVSIK PGVTQKVGEI ARTGSTPEVS TVDAMLDLIR NMFPENLVQA A FQQYKTKR ...String: GPMGKPARKG AEWKRFLKNN WVLLSTVAAV VLGITTGVLV REHSNLSTLE KFYFAFPGEI LMRMLKLIIL PLIISSMITG VAALDSNVS GKIGLRAVVY YFATTLIAVI LGIVLVVSIK PGVTQKVGEI ARTGSTPEVS TVDAMLDLIR NMFPENLVQA A FQQYKTKR EEVKPPSDPE MTMTEESFTA VMTTAISKTK TKEYKIVGMY SDGINVLGLI VFALVFGLVI GKMGEKGQIL VD FFNALSD ATMKIVQIIM WYMPLGILFL IAGCIIEVED WEIFRKLGLY MATVLTGLAI HSIVILPLIY FIVVRKNPFR FAM GMAQAL LTALMISSSS ATLPVTFRCA EENNQVDKRI TRFVLPVGAT INMDGTALYE AVAAVFIAQL NDLDLGIGQI ITIS ITATS ASIGAAGVPQ AGLVTMVIVL SAVGLPAEDV TLIIAVDCLL DRFRTMVNVL GDAFGTGIVE KLSKKELEQM DVSSE VNIV NPFALESTIL DNEDSDTKKS YVNGGFAVDK SDTISFTQTS QF UniProtKB: Excitatory amino acid transporter 3 |
-Macromolecule #2: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 2 / Number of copies: 3 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Macromolecule #3: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 3 / Number of copies: 9 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #4: MERCURY (II) ION
| Macromolecule | Name: MERCURY (II) ION / type: ligand / ID: 4 / Number of copies: 3 / Formula: HG |
|---|---|
| Molecular weight | Theoretical: 200.59 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 12 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Tris is used for adjusting pH | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot 3s. | |||||||||||||||
| Details | Human EAAT3 with glutamate bound |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.266 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Protocol: RIGID BODY FIT | ||||||
| Output model |  PDB-8ctc: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)