+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pseudomonas phage E217 small terminase (TerS) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phage small Terminase / decamer / oligomer / DNA BINDING PROTEIN | |||||||||
| Function / homology | Phage protein Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas phage vB_PaeM_E217 (virus) Pseudomonas phage vB_PaeM_E217 (virus) | |||||||||
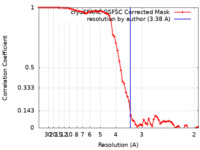
| Method | single particle reconstruction / cryo EM / Resolution: 3.38 Å | |||||||||
 Authors Authors | Lokareddy RK / Hou C-FD / Doll SG / Li F / Gillilan R / Forti F / Briani F / Cingolani G | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2022 Journal: J Mol Biol / Year: 2022Title: Terminase Subunits from the Pseudomonas-Phage E217. Authors: Ravi K Lokareddy / Chun-Feng David Hou / Steven G Doll / Fenglin Li / Richard E Gillilan / Francesca Forti / David S Horner / Federica Briani / Gino Cingolani /   Abstract: Pseudomonas phages are increasingly important biomedicines for phage therapy, but little is known about how these viruses package DNA. This paper explores the terminase subunits from the Myoviridae ...Pseudomonas phages are increasingly important biomedicines for phage therapy, but little is known about how these viruses package DNA. This paper explores the terminase subunits from the Myoviridae E217, a Pseudomonas-phage used in an experimental cocktail to eradicate P. aeruginosa in vitro and in animal models. We identified the large (TerL) and small (TerS) terminase subunits in two genes ∼58 kbs away from each other in the E217 genome. TerL presents a classical two-domain architecture, consisting of an N-terminal ATPase and C-terminal nuclease domain arranged into a bean-shaped tertiary structure. A 2.05 Å crystal structure of the C-terminal domain revealed an RNase H-like fold with two magnesium ions in the nuclease active site. Mutations in TerL residues involved in magnesium coordination had a dominant-negative effect on phage growth. However, the two ions identified in the active site were too far from each other to promote two-metal-ion catalysis, suggesting a conformational change is required for nuclease activity. We also determined a 3.38 Å cryo-EM reconstruction of E217 TerS that revealed a ring-like decamer, departing from the most common nonameric quaternary structure observed thus far. E217 TerS contains both N-terminal helix-turn-helix motifs enriched in basic residues and a central channel lined with basic residues large enough to accommodate double-stranded DNA. Overexpression of TerS caused a more than a 4-fold reduction of E217 burst size, suggesting a catalytic amount of the protein is required for packaging. Together, these data expand the molecular repertoire of viral terminase subunits to Pseudomonas-phages used for phage therapy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26858.map.gz emd_26858.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26858-v30.xml emd-26858-v30.xml emd-26858.xml emd-26858.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26858_fsc.xml emd_26858_fsc.xml | 6.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_26858.png emd_26858.png | 107.8 KB | ||
| Filedesc metadata |  emd-26858.cif.gz emd-26858.cif.gz | 5.1 KB | ||
| Others |  emd_26858_half_map_1.map.gz emd_26858_half_map_1.map.gz emd_26858_half_map_2.map.gz emd_26858_half_map_2.map.gz | 27.9 MB 28 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26858 http://ftp.pdbj.org/pub/emdb/structures/EMD-26858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26858 | HTTPS FTP |
-Validation report
| Summary document |  emd_26858_validation.pdf.gz emd_26858_validation.pdf.gz | 818.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26858_full_validation.pdf.gz emd_26858_full_validation.pdf.gz | 818.5 KB | Display | |
| Data in XML |  emd_26858_validation.xml.gz emd_26858_validation.xml.gz | 13.8 KB | Display | |
| Data in CIF |  emd_26858_validation.cif.gz emd_26858_validation.cif.gz | 17.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26858 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26858 | HTTPS FTP |
-Related structure data
| Related structure data |  7uxeMC  8dkrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26858.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26858.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_26858_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26858_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Small terminase decamer from Pseudomonas phage E217
| Entire | Name: Small terminase decamer from Pseudomonas phage E217 |
|---|---|
| Components |
|
-Supramolecule #1: Small terminase decamer from Pseudomonas phage E217
| Supramolecule | Name: Small terminase decamer from Pseudomonas phage E217 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage vB_PaeM_E217 (virus) Pseudomonas phage vB_PaeM_E217 (virus) |
| Molecular weight | Theoretical: 210 KDa |
-Macromolecule #1: Small terminase
| Macromolecule | Name: Small terminase / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage vB_PaeM_E217 (virus) Pseudomonas phage vB_PaeM_E217 (virus) |
| Molecular weight | Theoretical: 21.30468 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKFYSPDDL VTPQEFADPH FAAINQKRFD LYIDLRVQGY SSWRVFRAIW GEEHMDGPAQ ARIFAMESNP YYRKQFKAKL NATKTSDLW NPKTALHELL QMVRDPTVKD SSRLSAIKEL NVLAEITFVD ESGKTRIGRG LADFYASEAE AQTATVAAAA E ANSYVPEG EEGDFPSPTP EPTEEDRANP I UniProtKB: Phage protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7uxe: |
 Movie
Movie Controller
Controller




 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)