+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | IgA1 Protease with IgA1 substrate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationsecretory dimeric IgA immunoglobulin complex / monomeric IgA immunoglobulin complex / secretory IgA immunoglobulin complex / IgA immunoglobulin complex / glomerular filtration / IgG immunoglobulin complex / immunoglobulin complex, circulating / positive regulation of respiratory burst / complement activation, classical pathway / Scavenging of heme from plasma ...secretory dimeric IgA immunoglobulin complex / monomeric IgA immunoglobulin complex / secretory IgA immunoglobulin complex / IgA immunoglobulin complex / glomerular filtration / IgG immunoglobulin complex / immunoglobulin complex, circulating / positive regulation of respiratory burst / complement activation, classical pathway / Scavenging of heme from plasma / antigen binding / serine-type peptidase activity / Cell surface interactions at the vascular wall / B cell receptor signaling pathway / metalloendopeptidase activity / antibacterial humoral response / blood microparticle / adaptive immune response / immune response / extracellular space / extracellular exosome / extracellular region / zinc ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Gemella haemolysans (bacteria) / Gemella haemolysans (bacteria) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.56 Å | |||||||||
 Authors Authors | Eisenmesser ZE / Zheng H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: A substrate-induced gating mechanism is conserved among Gram-positive IgA1 metalloproteases. Authors: Jasmina S Redzic / Jeremy Rahkola / Norman Tran / Todd Holyoak / Eunjeong Lee / Antonio Javier Martín-Galiano / Nancy Meyer / Hongjin Zheng / Elan Eisenmesser /    Abstract: The mucosal adaptive immune response is dependent on the production of IgA antibodies and particularly IgA1, yet opportunistic bacteria have evolved mechanisms to specifically block this response by ...The mucosal adaptive immune response is dependent on the production of IgA antibodies and particularly IgA1, yet opportunistic bacteria have evolved mechanisms to specifically block this response by producing IgA1 proteases (IgA1Ps). Our lab was the first to describe the structures of a metal-dependent IgA1P (metallo-IgA1P) produced from Gram-positive Streptococcus pneumoniae both in the absence and presence of its IgA1 substrate through cryo-EM single particle reconstructions. This prior study revealed an active-site gating mechanism reliant on substrate-induced conformational changes to the enzyme that begged the question of whether such a mechanism is conserved among the wider Gram-positive metallo-IgA1P subfamily of virulence factors. Here, we used cryo-EM to characterize the metallo-IgA1P of a more distantly related family member from Gemella haemolysans, an emerging opportunistic pathogen implicated in meningitis, endocarditis, and more recently bacteremia in the elderly. While the substrate-free structures of these two metallo-IgA1Ps exhibit differences in the relative starting positions of the domain responsible for gating substrate, the enzymes have similar domain orientations when bound to IgA1. Together with biochemical studies that indicate these metallo-IgA1Ps have similar binding affinities and activities, these data indicate that metallo-IgA1P binding requires the specific IgA1 substrate to open the enzymes for access to their active site and thus, largely conform to an "induced fit" model. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26813.map.gz emd_26813.map.gz | 165.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26813-v30.xml emd-26813-v30.xml emd-26813.xml emd-26813.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26813.png emd_26813.png | 444.2 KB | ||
| Masks |  emd_26813_msk_1.map emd_26813_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26813.cif.gz emd-26813.cif.gz | 7.2 KB | ||
| Others |  emd_26813_half_map_1.map.gz emd_26813_half_map_1.map.gz emd_26813_half_map_2.map.gz emd_26813_half_map_2.map.gz | 301.5 MB 301.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26813 http://ftp.pdbj.org/pub/emdb/structures/EMD-26813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26813 | HTTPS FTP |
-Related structure data
| Related structure data |  7uvlMC  7uvkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26813.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26813.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.802 Å | ||||||||||||||||||||||||||||||||||||
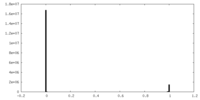
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26813_msk_1.map emd_26813_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
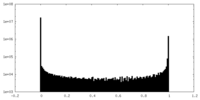
| Density Histograms |
-Half map: #2
| File | emd_26813_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
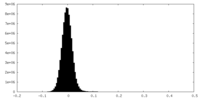
| Density Histograms |
-Half map: #1
| File | emd_26813_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
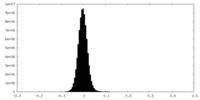
| Density Histograms |
- Sample components
Sample components
-Entire : IgA1 Protease with IgA1 substrate
| Entire | Name: IgA1 Protease with IgA1 substrate |
|---|---|
| Components |
|
-Supramolecule #1: IgA1 Protease with IgA1 substrate
| Supramolecule | Name: IgA1 Protease with IgA1 substrate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Gemella haemolysans (bacteria) Gemella haemolysans (bacteria) |
-Macromolecule #1: LPXTG-motif cell wall anchor domain protein
| Macromolecule | Name: LPXTG-motif cell wall anchor domain protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Gemella haemolysans (bacteria) Gemella haemolysans (bacteria) |
| Molecular weight | Theoretical: 146.820328 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DEKKDDIKEE KQVDKKLELR NISNVELYTL ENNKYRHVSS LSSVPTNPEA YFMKVKSENF KDVMLPVKSI ESARKDNQDV YKIVGQAND LIQHENNITL ENYTYYLPKT VNSENGVYTS FKNLVDAMNI NPYGTFRLGA TMDAREVELS DGQESYINKE F SGKLIGEN ...String: DEKKDDIKEE KQVDKKLELR NISNVELYTL ENNKYRHVSS LSSVPTNPEA YFMKVKSENF KDVMLPVKSI ESARKDNQDV YKIVGQAND LIQHENNITL ENYTYYLPKT VNSENGVYTS FKNLVDAMNI NPYGTFRLGA TMDAREVELS DGQESYINKE F SGKLIGEN KGKYYAIYNL KKPLFKALSH ATIQDLSIKE ANVSSKEDAA TIAKEAKNDT TIANVHSSGV IAGERSIGGL IS QVTDSTI SNSSFTGRIT NTYDTTATYQ IGGLVGKLSG VGALIEKSIS SIDMATNANT GDQVVGGVAG VVDKKATIRN SYV EGNLNN VKPFGKVGGV VGNLWDRETS EVSNSGNLTN VLSDVNVTNG NAIAGYDFNG IKATNTYSNK NNKVVKVVQV DDEV LSKDS EEQRGTVLEN NIVLEKKIEL VPKKNTKIED FNFSSRYETD YKNLKDADVS RLRVYKNIEK LLPFYNRETI VKYGN LVDA NNTLYTKDLV SVVPMKDKEV ISDINKNKTS INKLLLHYSD NTSQTLDIKY LQDFSKVAEY EIANTKLIYT PNTLLH SYN NIVKAVLNDL KSVQYDSDAV RKVLDISSNI KLTELYLDEQ FTKTKANIED SLSKLLSADA VIAENSNSII DNYVIEK IK NNKEALLLGL TYLERWYNFK YDNTSAKDLV LYHLDFFGKS NSSALDNVIE LGKSGFNNLL AKNNVITYNV LLSKNYGT E GLFKALEGYR KVFLPNVSNN DWFKTQSKAY IVEEKSTIPE VSSKQSKQGT EHSIGVYDRL TSPSWKYQSM VLPLLTLPE EKMIFMIANI STIGFGAYDR YRSSEYPKGD KLNRFVEENA QAAAKRFRDH YDYWYKILDK ENKEKLFRSV LVYDAFRFGN DTNKETQEA NFETNNPVIK NFFGPAGNNV VHNKHGAYAT GDAFYYMAYR MLDKSGAVTY THAMTHNSDR EIYLGGYGRR S GLGPEFYA KGLLQAPDHS YDPTITINSV LKYDDSENST RLQIADPTQR FTNVEDLHNY MHNMFDLIYT LEILEGRAVA KL DYNEKND LLRKIENIYK KDPDGNSVYA TNAVRRLTSD EIKNLTSFDK LIENDVITRR GYIDQGEYER NGYHTINLFS PIY SALSSK IGTPGDLMGR RMAFELLAAK GYKEGMVPYI SNQYEKEAKD RGSKIRSYGK EIGLVTDDLV LEKVFNKKYG SWVE FKKDM YKERVEQFSK LNRVSFFDPN GPWGRQKNVT VNNISVLEKM IETAVREDAE DFTAQVYPDT NSRVLKLKKA IFKAY LDQT KDFRTSIFGG K UniProtKB: LPXTG-motif cell wall anchor domain protein |
-Macromolecule #2: Immunoglobulin alpha-1 heavy constant
| Macromolecule | Name: Immunoglobulin alpha-1 heavy constant / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.784846 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CHPRLSLHRP ALEDLLLGSE ANLTCTLTGL RDASGVTFTW TPSSGKSAVQ GPPERDLCGC YSVSSVLPGC AEPWNHGKTF TCTAAYPES KTPLTATLSK SGNTFRPEVH LLPPPSEELA LNELVTLTCL ARGFSPKDVL VRWLQGSQEL PREKYLTWAS R QEPSQGTT ...String: CHPRLSLHRP ALEDLLLGSE ANLTCTLTGL RDASGVTFTW TPSSGKSAVQ GPPERDLCGC YSVSSVLPGC AEPWNHGKTF TCTAAYPES KTPLTATLSK SGNTFRPEVH LLPPPSEELA LNELVTLTCL ARGFSPKDVL VRWLQGSQEL PREKYLTWAS R QEPSQGTT TFAVTSILRV AAEDWKKGDT FSCMVGHEAL PLAFTQKTID R UniProtKB: Immunoglobulin heavy constant alpha 1 |
-Macromolecule #3: Immunoglobulin alpha-1 light chain
| Macromolecule | Name: Immunoglobulin alpha-1 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.009725 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVMTQSPLS LSVTPGEPAS ISCRSSQSLL RRDGHNDLEW YLQKPGQSPQ PLIYLGSTRA SGVPDRFSGS GSGTDFTLKI IRVEAEDAG TYYCMQNKQT PLTFGQGTRL EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV ...String: DIVMTQSPLS LSVTPGEPAS ISCRSSQSLL RRDGHNDLEW YLQKPGQSPQ PLIYLGSTRA SGVPDRFSGS GSGTDFTLKI IRVEAEDAG TYYCMQNKQT PLTFGQGTRL EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE C |
-Macromolecule #4: Immunoglobulin alpha-1 heavy chain
| Macromolecule | Name: Immunoglobulin alpha-1 heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.554428 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGGSLKL SCAASGFTLS GSNVHWVRQA SGKGLEWVGR IKRNAESDAT AYAASMRGRL TISRDDSKNT AFLQMNSLK SDDTAMYYCV IRGDVYNRQW GQGTLVTVSS ASPTSPKVFP LSLCSTQPDG NVVIACLVQG FFPQEPLSVT W SESGQGVT ...String: EVQLVESGGG LVQPGGSLKL SCAASGFTLS GSNVHWVRQA SGKGLEWVGR IKRNAESDAT AYAASMRGRL TISRDDSKNT AFLQMNSLK SDDTAMYYCV IRGDVYNRQW GQGTLVTVSS ASPTSPKVFP LSLCSTQPDG NVVIACLVQG FFPQEPLSVT W SESGQGVT ARNFPPSQDA SGDLYTTSSQ LTLPATQCLA GKSVTCHVKH YTNPSQDVTV PCPVPSTPPT PSPS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)