+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
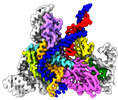
| Title | IscB and wRNA bound to Target DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / IscB / HEARO RNA / omega RNA / RNA BINDING PROTEIN-RNA-DNA complex | |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||
 Authors Authors | Schuler GA / Hu C / Ke A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structural basis for RNA-guided DNA cleavage by IscB-ωRNA and mechanistic comparison with Cas9. Authors: Gabriel Schuler / Chunyi Hu / Ailong Ke /  Abstract: Class 2 CRISPR effectors Cas9 and Cas12 may have evolved from nucleases in IS200/IS605 transposons. IscB is about two-fifths the size of Cas9 but shares a similar domain organization. The associated ...Class 2 CRISPR effectors Cas9 and Cas12 may have evolved from nucleases in IS200/IS605 transposons. IscB is about two-fifths the size of Cas9 but shares a similar domain organization. The associated ωRNA plays the combined role of CRISPR RNA (crRNA) and trans-activating CRISPR RNA tracrRNA) to guide double-stranded DNA (dsDNA) cleavage. Here we report a 2.78-angstrom cryo-electron microscopy structure of IscB-ωRNA bound to a dsDNA target, revealing the architectural and mechanistic similarities between IscB and Cas9 ribonucleoproteins. Target-adjacent motif recognition, R-loop formation, and DNA cleavage mechanisms are explained at high resolution. ωRNA plays the equivalent function of REC domains in Cas9 and contacts the RNA-DNA heteroduplex. The IscB-specific PLMP domain is dispensable for RNA-guided DNA cleavage. The transition from ancestral IscB to Cas9 involved dwarfing the ωRNA and introducing protein domain replacements. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26782.map.gz emd_26782.map.gz | 42.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26782-v30.xml emd-26782-v30.xml emd-26782.xml emd-26782.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26782.png emd_26782.png | 151.9 KB | ||
| Filedesc metadata |  emd-26782.cif.gz emd-26782.cif.gz | 6.5 KB | ||
| Others |  emd_26782_additional_1.map.gz emd_26782_additional_1.map.gz emd_26782_additional_2.map.gz emd_26782_additional_2.map.gz emd_26782_additional_3.map.gz emd_26782_additional_3.map.gz emd_26782_half_map_1.map.gz emd_26782_half_map_1.map.gz emd_26782_half_map_2.map.gz emd_26782_half_map_2.map.gz | 43.4 MB 41.8 MB 41.8 MB 77.5 MB 77.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26782 http://ftp.pdbj.org/pub/emdb/structures/EMD-26782 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26782 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26782 | HTTPS FTP |
-Related structure data
| Related structure data |  7utnMC  8cszC  8ctlC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26782.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26782.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
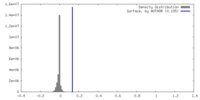
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #3
| File | emd_26782_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
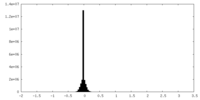
| Density Histograms |
-Additional map: #2
| File | emd_26782_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
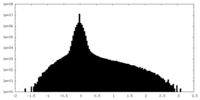
| Density Histograms |
-Additional map: #1
| File | emd_26782_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
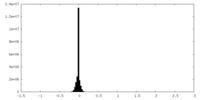
| Density Histograms |
-Half map: #2
| File | emd_26782_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26782_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of IscB in complex with RNA and target DNA
| Entire | Name: Cryo-EM structure of IscB in complex with RNA and target DNA |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of IscB in complex with RNA and target DNA
| Supramolecule | Name: Cryo-EM structure of IscB in complex with RNA and target DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 190 kDa/nm |
-Macromolecule #1: IscB
| Macromolecule | Name: IscB / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 56.688477 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAVVYVISKS GKPLMPTTRC GHVRILLKEG KARVVERKPF TIQLTYESAE ETQPLVLGID PGRTNIGMSV VTESGESVFN AQIETRNKD VPKLMKDRKQ YRMAHRRLKR RCKRRRRAKA AGTAFEEGEK QRLLPGCFKP ITCKSIRNKE ARFNNRKRPV G WLTPTANH ...String: MAVVYVISKS GKPLMPTTRC GHVRILLKEG KARVVERKPF TIQLTYESAE ETQPLVLGID PGRTNIGMSV VTESGESVFN AQIETRNKD VPKLMKDRKQ YRMAHRRLKR RCKRRRRAKA AGTAFEEGEK QRLLPGCFKP ITCKSIRNKE ARFNNRKRPV G WLTPTANH LLVTHLNVVK KVQKILPVAK VVLELNRFSF MAMNNPKVQR WQYQRGPLYG KGSVEEAVSM QQDGHCLFCK HG IDHYHHV VPRRKNGSET LENRVGLCEE HHRLVHTDKE WEANLASKKS GMNKKYHALS VLNQIIPYLA DQLADMFPGN FCV TSGQDT YLFREEHGIP KDHYLDAYCI ACSALTDAKK VSSPKGRPYM VHQFRRHDRQ ACHKANLNRS YYMGGKLVAT NRHK AMDQK TDSLEEYRAA HSAADVSKLT VKHPSAQYKD MSRIMPGSIL VSGEGKLFTL SRSEGRNKGQ VNYFVSTEGI KYWAR KCQY LRNNGGLQIY V |
-Macromolecule #2: RNA (222-MER)
| Macromolecule | Name: RNA (222-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 71.877797 KDa |
| Sequence | String: AAAAGAGUGA ACGAGAGGCU CUUCCAACUU UAUGGUUGCG ACCGUAGGUU GAAAGAGCAC AGGCUGAGAC AUUCGUAAGG CCGAAAGAC CGGACGCACC CUGGGAUUUC CCCAGUCCCC GGAACUGCAU AGCGGAUGCC AGUUGAUGGA GCAAUCUAUC A GAUAAGCC ...String: AAAAGAGUGA ACGAGAGGCU CUUCCAACUU UAUGGUUGCG ACCGUAGGUU GAAAGAGCAC AGGCUGAGAC AUUCGUAAGG CCGAAAGAC CGGACGCACC CUGGGAUUUC CCCAGUCCCC GGAACUGCAU AGCGGAUGCC AGUUGAUGGA GCAAUCUAUC A GAUAAGCC AGGGGGAACA AUCACCUCUC UGUAUCAGAG AGAGUUUUAC AAAAGGAGGA ACGG |
-Macromolecule #3: DNA target strand
| Macromolecule | Name: DNA target strand / type: dna / ID: 3 Details: phosphorothioate (PS) bonds * GCCACGGGCTGACCTCGACTTCTAGT*C*T*C*G*T*T*CACTCTTTTGCCGTACCCTCGTGGGGCG Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 18.305646 KDa |
| Sequence | String: (DG)(DC)(DC)(DA)(DC)(DG)(DG)(DG)(DC)(DT) (DG)(DA)(DC)(DC)(DT)(DC)(DG)(DA)(DC)(DT) (DT)(DC)(DT)(DA)(DG)(DT)(DC)(DT)(DC) (DG)(DT)(DT)(DC)(DA)(DC)(DT)(DC)(DT)(DT) (DT) (DT)(DG)(DC)(DC)(DG)(DT) ...String: (DG)(DC)(DC)(DA)(DC)(DG)(DG)(DG)(DC)(DT) (DG)(DA)(DC)(DC)(DT)(DC)(DG)(DA)(DC)(DT) (DT)(DC)(DT)(DA)(DG)(DT)(DC)(DT)(DC) (DG)(DT)(DT)(DC)(DA)(DC)(DT)(DC)(DT)(DT) (DT) (DT)(DG)(DC)(DC)(DG)(DT)(DA)(DC) (DC)(DC)(DT)(DC)(DG)(DT)(DG)(DG)(DG)(DG) (DC)(DC) |
-Macromolecule #4: DNA non-target strand
| Macromolecule | Name: DNA non-target strand / type: dna / ID: 4 Details: phosphorothioate (PS) bond * CGCCCCACGAGGGTACGGCAAAAGA*G*T*T*T*T*T*TTTACTAGAAGTCGAGGTCAGCCCGTGGC Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 18.557873 KDa |
| Sequence | String: (DC)(DG)(DC)(DC)(DC)(DC)(DA)(DC)(DG)(DA) (DG)(DG)(DG)(DT)(DA)(DC)(DG)(DG)(DC)(DA) (DA)(DA)(DA)(DG)(DA)(DG)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DA)(DC)(DT)(DA)(DG) (DA) (DA)(DG)(DT)(DC)(DG)(DA) ...String: (DC)(DG)(DC)(DC)(DC)(DC)(DA)(DC)(DG)(DA) (DG)(DG)(DG)(DT)(DA)(DC)(DG)(DG)(DC)(DA) (DA)(DA)(DA)(DG)(DA)(DG)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DA)(DC)(DT)(DA)(DG) (DA) (DA)(DG)(DT)(DC)(DG)(DA)(DG)(DG) (DT)(DC)(DA)(DG)(DC)(DC)(DC)(DG)(DT)(DG) (DG)(DC) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.25 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 100 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Pressure: 0.039 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 6.5 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.74 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 159201 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)