[English] 日本語
 Yorodumi
Yorodumi- EMDB-26622: The V1 region of bovine V-ATPase in complex with human mEAK7 (foc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The V1 region of bovine V-ATPase in complex with human mEAK7 (focused refinement) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | proton transport / mTOR signaling | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationROS and RNS production in phagocytes / Insulin receptor recycling / Transferrin endocytosis and recycling / Amino acids regulate mTORC1 / Ion channel transport / pH reduction / synaptic vesicle lumen acidification / cellular response to increased oxygen levels / vacuolar proton-transporting V-type ATPase, V1 domain / clathrin-coated vesicle membrane ...ROS and RNS production in phagocytes / Insulin receptor recycling / Transferrin endocytosis and recycling / Amino acids regulate mTORC1 / Ion channel transport / pH reduction / synaptic vesicle lumen acidification / cellular response to increased oxygen levels / vacuolar proton-transporting V-type ATPase, V1 domain / clathrin-coated vesicle membrane / proton-transporting V-type ATPase complex / cell projection organization / vacuolar acidification / Neutrophil degranulation / proton-transporting ATPase activity, rotational mechanism / ATP metabolic process / H+-transporting two-sector ATPase / transport vesicle / proton transmembrane transport / synaptic vesicle membrane / melanosome / intracellular iron ion homeostasis / lysosome / endosome / cilium / apical plasma membrane / lysosomal membrane / ATP hydrolysis activity / ATP binding / plasma membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
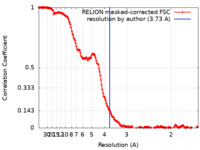
| Method | single particle reconstruction / cryo EM / Resolution: 3.73 Å | ||||||||||||
 Authors Authors | Wang R / Li X | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Molecular basis of mEAK7-mediated human V-ATPase regulation. Authors: Rong Wang / Yu Qin / Xiao-Song Xie / Xiaochun Li /  Abstract: The activity of V-ATPase is well-known to be regulated by reversible dissociation of its V and V domains in response to growth factor stimulation, nutrient sensing, and cellular differentiation. The ...The activity of V-ATPase is well-known to be regulated by reversible dissociation of its V and V domains in response to growth factor stimulation, nutrient sensing, and cellular differentiation. The molecular basis of its regulation by an endogenous modulator without affecting V-ATPase assembly remains unclear. Here, we discover that a lysosome-anchored protein termed (mammalian Enhancer-of-Akt-1-7 (mEAK7)) binds to intact V-ATPase. We determine cryo-EM structure of human mEAK7 in complex with human V-ATPase in native lipid-containing nanodiscs. The structure reveals that the TLDc domain of mEAK7 engages with subunits A, B, and E, while its C-terminal domain binds to subunit D, presumably blocking V-V torque transmission. Our functional studies suggest that mEAK7, which may act as a V-ATPase inhibitor, does not affect the activity of V-ATPase in vitro. However, overexpression of mEAK7 in HCT116 cells that stably express subunit a4 of V-ATPase represses the phosphorylation of ribosomal protein S6. Thus, this finding suggests that mEAK7 potentially links mTOR signaling with V-ATPase activity. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26622.map.gz emd_26622.map.gz | 394.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26622-v30.xml emd-26622-v30.xml emd-26622.xml emd-26622.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26622_fsc.xml emd_26622_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_26622.png emd_26622.png | 80.5 KB | ||
| Filedesc metadata |  emd-26622.cif.gz emd-26622.cif.gz | 6.8 KB | ||
| Others |  emd_26622_half_map_1.map.gz emd_26622_half_map_1.map.gz emd_26622_half_map_2.map.gz emd_26622_half_map_2.map.gz | 337.9 MB 338.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26622 http://ftp.pdbj.org/pub/emdb/structures/EMD-26622 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26622 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26622 | HTTPS FTP |
-Related structure data
| Related structure data |  7uneMC  7unfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26622.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26622.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_26622_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
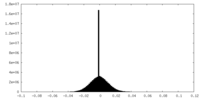
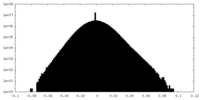
| Density Histograms |
-Half map: #2
| File | emd_26622_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the V1 region of bovine V-ATPase in complex ...
| Entire | Name: Cryo-EM structure of the V1 region of bovine V-ATPase in complex with human mEAK7 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the V1 region of bovine V-ATPase in complex ...
| Supramolecule | Name: Cryo-EM structure of the V1 region of bovine V-ATPase in complex with human mEAK7 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: V-type proton ATPase catalytic subunit A
| Macromolecule | Name: V-type proton ATPase catalytic subunit A / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: H+-transporting two-sector ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 68.420914 KDa |
| Sequence | String: MDFSKLPKIR DEDKESTFGY VHGVSGPVVT ACDMAGAAMY ELVRVGHSEL VGEIIRLEGD MATIQVYEET SGVSVGDPVL RTGKPLSVE LGPGIMGAIF DGIQRPLSDI SSQTQSIYIP RGVNVSALSR DVKWDFTPCK NLRVGSHITG GDIYGIVNEN S LIKHKIML ...String: MDFSKLPKIR DEDKESTFGY VHGVSGPVVT ACDMAGAAMY ELVRVGHSEL VGEIIRLEGD MATIQVYEET SGVSVGDPVL RTGKPLSVE LGPGIMGAIF DGIQRPLSDI SSQTQSIYIP RGVNVSALSR DVKWDFTPCK NLRVGSHITG GDIYGIVNEN S LIKHKIML PPRNRGTVTY IAPPGNYDTS DVVLELEFEG IKEKFSMVQV WPVRQVRPVT EKLPANHPLL TGQRVLDALF PC VQGGTTA IPGAFGCGKT VISQSLSKYS NSDVIIYVGC GERGNEMSEV LRDFPELTME VDGKVESIMK RTALVANTSN MPV AAREAS IYTGITLSEY FRDMGYHVSM MADSTSRWAE ALREISGRLA EMPADSGYPA YLGARLASFY ERAGRVKCLG NPER EGSVS IVGAVSPPGG DFSDPVTSAT LGIVQVFWGL DKKLAQRKHF PSVNWLISYS KYMRALDEYY DKHFTEFVPL RTKAK EILQ EEEDLAEIVQ LVGKASLAET DKITLEVAKL IKDDFLQQNG YTPYDRFCPF YKTVGMLSNM IAFYDMARRA VETTAQ SDN KITWSIIREH MGEILYKLSS MKFKDPVKDG EAKIKADYAQ LLEDMQNAFR SLED UniProtKB: V-type proton ATPase catalytic subunit A |
-Macromolecule #2: V-type proton ATPase subunit D
| Macromolecule | Name: V-type proton ATPase subunit D / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.297893 KDa |
| Sequence | String: MSGKDRIEIF PSRMAQTIMK ARLKGAQTGR NLLKKKSDAL TLRFRQILKK IIETKMLMGE VMREAAFSLA EAKFTAGDFS TTVIQNVNK AQVKIRAKKD NVAGVTLPVF EHYHEGTDSY ELTGLARGGE QLAKLKRNYA KAVELLVELA SLQTSFVTLD E AIKITNRR ...String: MSGKDRIEIF PSRMAQTIMK ARLKGAQTGR NLLKKKSDAL TLRFRQILKK IIETKMLMGE VMREAAFSLA EAKFTAGDFS TTVIQNVNK AQVKIRAKKD NVAGVTLPVF EHYHEGTDSY ELTGLARGGE QLAKLKRNYA KAVELLVELA SLQTSFVTLD E AIKITNRR VNAIEHVIIP RIERTLAYII TELDEREREE FYRLKKIQEK KKILKEKSDK DLEQRRAAGE VIEPANLLAE EK DEDLLFE UniProtKB: V-type proton ATPase subunit D |
-Macromolecule #3: KIAA1609 protein, isoform CRA_a
| Macromolecule | Name: KIAA1609 protein, isoform CRA_a / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.982465 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHGN SRSRVGRSFC SQFLPEEQAE IDQLFDALSS DKNSPNVSSK SFSLKALQNH VGEALPPEMV TRLYDGMRRV DLTGKAKGP SENVSQEQFT ASMSHLLKGN SEEKSLMIMK MISATEGPVK AREVQKFTED LVGSVVHVLS HRQELRGWTG K EAPGPNPR ...String: MGHHHHHHGN SRSRVGRSFC SQFLPEEQAE IDQLFDALSS DKNSPNVSSK SFSLKALQNH VGEALPPEMV TRLYDGMRRV DLTGKAKGP SENVSQEQFT ASMSHLLKGN SEEKSLMIMK MISATEGPVK AREVQKFTED LVGSVVHVLS HRQELRGWTG K EAPGPNPR VQVLAAQLLS DMKLQDGKRL LGPQWLDYDC DRAVIEDWVF RVPHVAIFLS VVICKGFLVL CSSLDLTTLV PE RQVDQGR GFESILDVLS VMYINAQLPR EQRHRWRLLF SSELHGHSFS QLCGHITHRG PCVAVLEDHD KHVFGGFASC SWE VKPQFQ GDNRCFLFSI CPSMAVYTHT GYNDHYMYLN HGQQTIPNGL GMGGQHNYFG LWVDVDFGKG HSRAKPTCTT YNSP QLSAQ ENFQFDKMEV WAVGDPSEEQ LAKGNKSILD ADPEAQALLE ISGHSRHSEG LREVPDDE UniProtKB: KIAA1609 protein, isoform CRA_a |
-Macromolecule #4: V-type proton ATPase subunit E 1
| Macromolecule | Name: V-type proton ATPase subunit E 1 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.178371 KDa |
| Sequence | String: MALSDADVQK QIKHMMAFIE QEANEKAEEI DAKAEEEFNI EKGRLVQTQR LKIMEYYEKK EKQIEQQKKI QMSNLMNQAR LKVLRARDD LITDLLNEAK QRLSKVVKDT TRYQVLLDGL VLQGLYQLLE PRMIVRCRKQ DFPLVKAAVQ KAIPVYKVAT K RDVDVQID ...String: MALSDADVQK QIKHMMAFIE QEANEKAEEI DAKAEEEFNI EKGRLVQTQR LKIMEYYEKK EKQIEQQKKI QMSNLMNQAR LKVLRARDD LITDLLNEAK QRLSKVVKDT TRYQVLLDGL VLQGLYQLLE PRMIVRCRKQ DFPLVKAAVQ KAIPVYKVAT K RDVDVQID QEAYLPEEIA GGVEIYNGDR KIKVSNTLES RLDLIAQQMM PEVRGALFGA NANRKFLD UniProtKB: V-type proton ATPase subunit E 1 |
-Macromolecule #5: V-type proton ATPase subunit B, brain isoform
| Macromolecule | Name: V-type proton ATPase subunit B, brain isoform / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.637555 KDa |
| Sequence | String: MALRAMRGIV NGAAPELPVP TSGPLAGSRE QALAVSRNYL SQPRLTYKTV SGVNGPLVIL DHVKFPRYAE IVHLTLPDGT KRSGQVLEV SGSKAVVQVF EGTSGIDAKK TSCEFTGDIL RTPVSEDMLG RVFNGSGKPI DRGPVVLAED FLDIMGQPIN P QCRIYPEE ...String: MALRAMRGIV NGAAPELPVP TSGPLAGSRE QALAVSRNYL SQPRLTYKTV SGVNGPLVIL DHVKFPRYAE IVHLTLPDGT KRSGQVLEV SGSKAVVQVF EGTSGIDAKK TSCEFTGDIL RTPVSEDMLG RVFNGSGKPI DRGPVVLAED FLDIMGQPIN P QCRIYPEE MIQTGISAID GMNSIARGQK IPIFSAAGLP HNEIAAQICR QAGLVKKSKD VVDYSEENFA IVFAAMGVNM ET ARFFKSD FEENGSMDNV CLFLNLANDP TIERIITPRL ALTTAEFLAY QCEKHVLVIL TDMSSYAEAL REVSAAREEV PGR RGFPGY MYTDLATIYE RAGRVEGRNG SITQIPILTM PNDDITHPIP DLTGYITEGQ IYVDRQLHNR QIYPPINVLP SLSR LMKSA IGEGMTRKDH ADVSNQLYAC YAIGKDVQAM KAVVGEEALT SDDLLYLEFL QKFERNFIAQ GPYENRTVYE TLDIG WQLL RIFPKEMLKR IPQSTLSEFY PRDSAKH UniProtKB: V-type proton ATPase subunit B, brain isoform |
-Macromolecule #6: V-type proton ATPase subunit G
| Macromolecule | Name: V-type proton ATPase subunit G / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.588344 KDa |
| Sequence | String: MASQSQGIQQ LLQAEKRAAE KVADARKRKA RRLKQAKEEA QMEVDQYRRE REQEFQSKQQ AAMGSQGNLS AEVEQATRRQ VQGMQSSQQ RNRERVLAQL LGMVCDVRPQ VHPNYRIAA UniProtKB: V-type proton ATPase subunit G 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)