+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure of Inactive MOR Bound to Alvimopan and Mb6 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Antagonist / Complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationOpioid Signalling / Peptide ligand-binding receptors / beta-endorphin receptor activity / morphine receptor activity / negative regulation of Wnt protein secretion / G-protein activation / G protein-coupled opioid receptor activity / G protein-coupled opioid receptor signaling pathway / G alpha (i) signalling events / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway ...Opioid Signalling / Peptide ligand-binding receptors / beta-endorphin receptor activity / morphine receptor activity / negative regulation of Wnt protein secretion / G-protein activation / G protein-coupled opioid receptor activity / G protein-coupled opioid receptor signaling pathway / G alpha (i) signalling events / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / negative regulation of nitric oxide biosynthetic process / regulation of NMDA receptor activity / positive regulation of neurogenesis / negative regulation of cytosolic calcium ion concentration / transmission of nerve impulse / G-protein alpha-subunit binding / sensory perception of pain / presynaptic modulation of chemical synaptic transmission / locomotory behavior / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / adenylate cyclase-activating dopamine receptor signaling pathway / presynapse / perikaryon / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of ERK1 and ERK2 cascade / endosome / axon / dendrite / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Robertson MJ / Skiniotis G | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structure determination of inactive-state GPCRs with a universal nanobody. Authors: Michael J Robertson / Makaía M Papasergi-Scott / Feng He / Alpay B Seven / Justin G Meyerowitz / Ouliana Panova / Maria Claudia Peroto / Tao Che / Georgios Skiniotis /  Abstract: Cryogenic electron microscopy (cryo-EM) has widened the field of structure-based drug discovery by allowing for routine determination of membrane protein structures previously intractable. Despite ...Cryogenic electron microscopy (cryo-EM) has widened the field of structure-based drug discovery by allowing for routine determination of membrane protein structures previously intractable. Despite representing one of the largest classes of therapeutic targets, most inactive-state G protein-coupled receptors (GPCRs) have remained inaccessible for cryo-EM because their small size and membrane-embedded nature impedes projection alignment for high-resolution map reconstructions. Here we demonstrate that the same single-chain camelid antibody (nanobody) recognizing a grafted intracellular loop can be used to obtain cryo-EM structures of inactive-state GPCRs at resolutions comparable or better than those obtained by X-ray crystallography. Using this approach, we obtained structures of neurotensin 1 receptor bound to antagonist SR48692, μ-opioid receptor bound to alvimopan, apo somatostatin receptor 2 and histamine receptor 2 bound to famotidine. We expect this rapid, straightforward approach to facilitate the broad exploration of GPCR inactive states without the need for extensive engineering and crystallization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26591.map.gz emd_26591.map.gz | 107.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26591-v30.xml emd-26591-v30.xml emd-26591.xml emd-26591.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26591.png emd_26591.png | 92 KB | ||
| Filedesc metadata |  emd-26591.cif.gz emd-26591.cif.gz | 6.1 KB | ||
| Others |  emd_26591_additional_1.map.gz emd_26591_additional_1.map.gz emd_26591_half_map_1.map.gz emd_26591_half_map_1.map.gz emd_26591_half_map_2.map.gz emd_26591_half_map_2.map.gz | 1.5 MB 116.2 MB 116.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26591 http://ftp.pdbj.org/pub/emdb/structures/EMD-26591 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26591 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26591 | HTTPS FTP |
-Validation report
| Summary document |  emd_26591_validation.pdf.gz emd_26591_validation.pdf.gz | 935.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26591_full_validation.pdf.gz emd_26591_full_validation.pdf.gz | 934.9 KB | Display | |
| Data in XML |  emd_26591_validation.xml.gz emd_26591_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_26591_validation.cif.gz emd_26591_validation.cif.gz | 16.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26591 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26591 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26591 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26591 | HTTPS FTP |
-Related structure data
| Related structure data |  7ul4MC  7ul2C  7ul3C  7ul5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26591.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26591.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8677 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Phenix model-free density modified map
| File | emd_26591_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Phenix model-free density modified map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26591_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26591_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of MOR and Nb6
| Entire | Name: Complex of MOR and Nb6 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of MOR and Nb6
| Supramolecule | Name: Complex of MOR and Nb6 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: MOR
| Supramolecule | Name: MOR / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Mb6
| Supramolecule | Name: Mb6 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Mu-type opioid receptor
| Macromolecule | Name: Mu-type opioid receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.920734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAMGPGNI SDCSDPLAPA SCSPAPGSWL NLSHVDGNQS DPCGPNRTGL GGSHSLCPQT GSPSMVTAI TIMALYSIVC VVGLFGNFLV MYVIVRYTKM KTATNIYIFN LALADALATS TLPFQSVNYL MGTWPFGNIL C KIVISIDY ...String: MKTIIALSYI FCLVFADYKD DDDAMGPGNI SDCSDPLAPA SCSPAPGSWL NLSHVDGNQS DPCGPNRTGL GGSHSLCPQT GSPSMVTAI TIMALYSIVC VVGLFGNFLV MYVIVRYTKM KTATNIYIFN LALADALATS TLPFQSVNYL MGTWPFGNIL C KIVISIDY YNMFTSIFTL CTMSVDRYIA VCHPVKALDF RTPRNAKIVN VCNWILSSAI GLPVMFMATT KYRQGSIDCT LT FSHPTWY WENLLKICVF IFAFIMPVLI ITVCYGLMIL RLKSVRLLSG SREKDRNLRR ITRMVLVVVA VFIVCWTPIH IYV IIKALI TIPETTFQTV SWHFCIALGY TNSCLNPVLY AFLDENFKRC FREFCIPTSS TIEQQNSARI RQNTREHPST ANTV DRTNH QLENLEAETA PLPDIHHHHH H UniProtKB: Mu-type opioid receptor |
-Macromolecule #2: Megabody 6
| Macromolecule | Name: Megabody 6 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 55.662078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVRKTTTSVI DTTNDAQNLL TQAQTIVNTL KDYCPILIAK SSSSNGGTNN ANTPSWQTAG GGKNSCATFG AEFSAASDM INNAQKIVQE TQQLSANQPK NITQPHNLNL NSPSSLTALA QKMLKNAQSQ AEILKLANQV ESDFNKLSSG H LKDYIGKC ...String: QVQLQESGGG LVRKTTTSVI DTTNDAQNLL TQAQTIVNTL KDYCPILIAK SSSSNGGTNN ANTPSWQTAG GGKNSCATFG AEFSAASDM INNAQKIVQE TQQLSANQPK NITQPHNLNL NSPSSLTALA QKMLKNAQSQ AEILKLANQV ESDFNKLSSG H LKDYIGKC DASAISSANM TMQNQKNNWG NGCAGVEETQ SLLKTSAADF NNQTPQINQA QNLANTLIQE LGNNTYEQLS RL LTNDNGT NSKTSAQAIN QAVNNLNERA KTLAGGTTNS PAYQATLLAL RSVLGLWNSM GYAVICGGYT KSPGENNQKD FHY TDENGN GTTINCGGST NSNGTHSYNG TNTLKADKNV SLSIEQYEKI HEAYQILSKA LKQAGLAPLN SKGEKLEAHV TTSK PSLRL SCAASGTIFR LYDMGWYRRV SGNQRELVAS ITSGGSTKYG DSVKGRFTIS RDNAKNTVYL QMSSLKPEDT AVYYC NAEY RTGIWEELLD GWGQGTQVTV SSHHHHHHEP EA |
-Macromolecule #3: N-[(2S)-2-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-y...
| Macromolecule | Name: N-[(2S)-2-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]methyl}-3-phenylpropanoyl]glycine type: ligand / ID: 3 / Number of copies: 1 / Formula: NG0 |
|---|---|
| Molecular weight | Theoretical: 424.533 Da |
| Chemical component information | 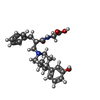 ChemComp-NG0: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 61.38 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 301332 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































