+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of AAV-PHP.eB, C1 Symmetry | |||||||||
 Map data Map data | Symmetry-free AAV-PHP.eB | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | gene therapy / AAV / capsid / blood-brain barrier / directed evolution / VIRUS | |||||||||
| Biological species |   Adeno-associated virus Adeno-associated virus | |||||||||
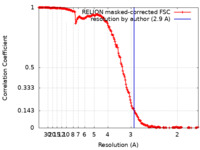
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Jang S / Shen HK / Ding X / Miles TF / Gradinaru V | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Ther Methods Clin Dev / Year: 2022 Journal: Mol Ther Methods Clin Dev / Year: 2022Title: Structural basis of receptor usage by the engineered capsid AAV-PHP.eB. Authors: Seongmin Jang / Hao K Shen / Xiaozhe Ding / Timothy F Miles / Viviana Gradinaru /  Abstract: Adeno-associated virus serotype 9 (AAV9) is a promising gene therapy vector for treating neurodegenerative diseases due to its ability to penetrate the blood-brain barrier. PHP.eB was engineered ...Adeno-associated virus serotype 9 (AAV9) is a promising gene therapy vector for treating neurodegenerative diseases due to its ability to penetrate the blood-brain barrier. PHP.eB was engineered from AAV9 by insertion of a 7-amino acid peptide and point mutation of neighboring residues, thereby enhancing potency in the central nervous system. Here, we report a 2.24-Å resolution cryo-electron microscopy structure of PHP.eB, revealing conformational differences from other 7-mer insertion capsid variants. In PHP.eB, the 7-mer loop adopts a bent conformation, mediated by an interaction between engineered lysine and aspartate residues. Further, we identify PKD2 as the main AAV receptor (AAVR) domain recognizing both AAV9 and PHP.eB and find that the PHP.eB 7-mer partially destabilizes this interaction. Analysis of previously reported AAV structures together with our pull-down data demonstrate that the 7-mer topology determined by the lysine-aspartate interaction dictates AAVR binding strength. Our results suggest that PHP.eB's altered tropism may arise from both an additional interaction with LY6A and weakening of its AAVR interaction. Changing the insertion length, but not sequence, modifies PKD2 binding affinity, suggesting that a steric clash impedes AAVR binding. This research suggests improved library designs for future AAV selections to identify non-LY6A-dependent vectors and modulate AAVR interaction strength. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26417.map.gz emd_26417.map.gz | 440.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26417-v30.xml emd-26417-v30.xml emd-26417.xml emd-26417.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26417_fsc.xml emd_26417_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_26417.png emd_26417.png | 214 KB | ||
| Filedesc metadata |  emd-26417.cif.gz emd-26417.cif.gz | 5 KB | ||
| Others |  emd_26417_half_map_1.map.gz emd_26417_half_map_1.map.gz emd_26417_half_map_2.map.gz emd_26417_half_map_2.map.gz | 383.6 MB 383.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26417 http://ftp.pdbj.org/pub/emdb/structures/EMD-26417 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26417 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26417 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26417.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26417.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Symmetry-free AAV-PHP.eB | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.869 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Symmetry-free AAV-PHP.eB
| File | emd_26417_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Symmetry-free AAV-PHP.eB | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Symmetry-free AAV-PHP.eB
| File | emd_26417_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Symmetry-free AAV-PHP.eB | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Adeno-associated virus
| Entire | Name:   Adeno-associated virus Adeno-associated virus |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus
| Supramolecule | Name: Adeno-associated virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: AAV-PHP.eB: Engineered AAV from AAV9 / NCBI-ID: 272636 / Sci species name: Adeno-associated virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:   Adeno-associated virus Adeno-associated virus |
| Sequence | String: MAADGYLPDW LEDNLSEGIR EWWALKPGAP QPKANQQHQD NARGLVLPGY KYLGPGNGLD KGEPVNAADA AALEHDKAYD QQLKAGDNPY LKYNHADAEF QERLKEDTSF GGNLGRAVFQ AKKRLLEPLG LVEEAAKTAP GKKRPVEQSP QEPDSSAGIG KSGAQPAKKR ...String: MAADGYLPDW LEDNLSEGIR EWWALKPGAP QPKANQQHQD NARGLVLPGY KYLGPGNGLD KGEPVNAADA AALEHDKAYD QQLKAGDNPY LKYNHADAEF QERLKEDTSF GGNLGRAVFQ AKKRLLEPLG LVEEAAKTAP GKKRPVEQSP QEPDSSAGIG KSGAQPAKKR LNFGQTGDTE SVPDPQPIGE PPAAPSGVGS LTMASGGGAP VADNNEGADG VGSSSGNWHC DSQWLGDRVI TTSTRTWALP TYNNHLYKQI SNSTSGGSSN DNAYFGYSTP WGYFDFNRFH CHFSPRDWQR LINNNWGFRP KRLNFKLFNI QVKEVTDNNG VKTIANNLTS TVQVFTDSDY QLPYVLGSAH EGCLPPFPAD VFMIPQYGYL TLNDGSQAVG RSSFYCLEYF PSQMLRTGNN FQFSYEFENV PFHSSYAHSQ SLDRLMNPLI DQYLYYLSKT INGSGQNQQT LKFSVAGPSN MAVQGRNYIP GPSYRQQRVS TTVTQNNNSE FAWPGASSWA LNGRNSLMNP GPAMASHKEG EDRFFPLSGS LIFGKQGTGR DNVDADKVMI TNEEEIKTTN PVATESYGQV ATNHQSDGTL AVPFKAQAQT GWVQNQGILP GMVWQDRDVY LQGPIWAKIP HTDGNFHPSP LMGGFGMKHP PPQILIKNTP VPADPPTAFN KDKLNSFITQ YSTGQVSVEI EWELQKENSK RWNPEIQYTS NYYKSNNVEF AVNTEGVYSE PRPIGTRYLT RNL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)