+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of DNPEP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNPEP / aminopeptidase / tetrahedral Symmetry / PEPTIDE BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationaspartyl aminopeptidase / aminopeptidase activity / metallopeptidase activity / proteolysis / zinc ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
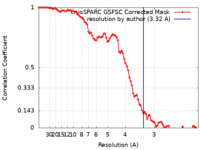
| Method | single particle reconstruction / cryo EM / Resolution: 3.32 Å | |||||||||
 Authors Authors | Morgan CE / Yu EW / Zhang Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Toward structural-omics of the bovine retinal pigment epithelium. Authors: Christopher E Morgan / Zhemin Zhang / Masaru Miyagi / Marcin Golczak / Edward W Yu /  Abstract: The use of an integrated systems biology approach to investigate tissues and organs has been thought to be impracticable in the field of structural biology, where the techniques mainly focus on ...The use of an integrated systems biology approach to investigate tissues and organs has been thought to be impracticable in the field of structural biology, where the techniques mainly focus on determining the structure of a particular biomacromolecule of interest. Here, we report the use of cryoelectron microscopy (cryo-EM) to define the composition of a raw bovine retinal pigment epithelium (RPE) lysate. From this sample, we simultaneously identify and solve cryo-EM structures of seven different RPE enzymes whose functions affect neurotransmitter recycling, iron metabolism, gluconeogenesis, glycolysis, axonal development, and energy homeostasis. Interestingly, dysfunction of these important proteins has been directly linked to several neurodegenerative disorders, including Huntington's disease, amyotrophic lateral sclerosis (ALS), Parkinson's disease, Alzheimer's disease, and schizophrenia. Our work underscores the importance of cryo-EM in facilitating tissue and organ proteomics at the atomic level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26350.map.gz emd_26350.map.gz | 14.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26350-v30.xml emd-26350-v30.xml emd-26350.xml emd-26350.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26350_fsc.xml emd_26350_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_26350.png emd_26350.png | 145.5 KB | ||
| Filedesc metadata |  emd-26350.cif.gz emd-26350.cif.gz | 5.9 KB | ||
| Others |  emd_26350_additional_1.map.gz emd_26350_additional_1.map.gz emd_26350_half_map_1.map.gz emd_26350_half_map_1.map.gz emd_26350_half_map_2.map.gz emd_26350_half_map_2.map.gz | 32.3 MB 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26350 http://ftp.pdbj.org/pub/emdb/structures/EMD-26350 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26350 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26350 | HTTPS FTP |
-Validation report
| Summary document |  emd_26350_validation.pdf.gz emd_26350_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26350_full_validation.pdf.gz emd_26350_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_26350_validation.xml.gz emd_26350_validation.xml.gz | 16.7 KB | Display | |
| Data in CIF |  emd_26350_validation.cif.gz emd_26350_validation.cif.gz | 21.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26350 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26350 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26350 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26350 | HTTPS FTP |
-Related structure data
| Related structure data |  7u5hMC  7u5iC  7u5jC  7u5kC  7u5lC  7u5mC  7u5nC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26350.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26350.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_26350_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
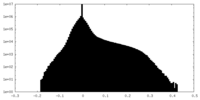
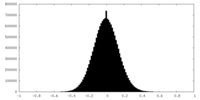
| Density Histograms |
-Half map: #2
| File | emd_26350_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
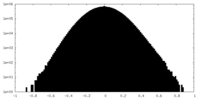
| Density Histograms |
-Half map: #1
| File | emd_26350_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
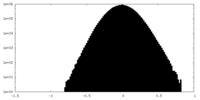
| Density Histograms |
- Sample components
Sample components
-Entire : Dodecamer of Aspartyl Aminopeptidase
| Entire | Name: Dodecamer of Aspartyl Aminopeptidase |
|---|---|
| Components |
|
-Supramolecule #1: Dodecamer of Aspartyl Aminopeptidase
| Supramolecule | Name: Dodecamer of Aspartyl Aminopeptidase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Aspartyl aminopeptidase
| Macromolecule | Name: Aspartyl aminopeptidase / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO / EC number: aspartyl aminopeptidase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.893188 KDa |
| Sequence | String: MSGRARKEAV QAAARELLKF VNRSPSPFHA VAECRSRLLQ AGFHELKETE SWDIKPESKY FLTRNSSTII AFAVGGQYVP GNGFSLIGA HTDSPCLRVK RRSRRSQVGF QQVGVETYGG GIWSTWFDRD LTLAGRVIVK CPTSGRLEQR LVHVDRPILR I PHLAIHLQ ...String: MSGRARKEAV QAAARELLKF VNRSPSPFHA VAECRSRLLQ AGFHELKETE SWDIKPESKY FLTRNSSTII AFAVGGQYVP GNGFSLIGA HTDSPCLRVK RRSRRSQVGF QQVGVETYGG GIWSTWFDRD LTLAGRVIVK CPTSGRLEQR LVHVDRPILR I PHLAIHLQ RNVNENFGPN MEMHLVPILA TSIQEELEKG TPEPGPLNAT DERHHSVLTS LLCAHLGLSP EDILEMELCL AD TQPAVLG GAYEEFIFAP RLDNLHSCFC ALQALIDSCS APASLAADPH VRMIALYDNE EVGSESAQGA QSLLTELVLR RIS ASPQHL TAFEEAIPKS YMISADMAHA VHPNYLDKHE ENHRPLFHKG PVIKVNSKQR YASNAVSEAL IREVASSVGV PLQD LMVRN DSPCGTTIGP ILASRLGLRV LDLGSPQLAM HSIRETACTT GVLQTITLFK GFFELFPSLS RSLLVD UniProtKB: Aspartyl aminopeptidase |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 24 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 37.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)