+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
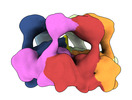
| Title | Skd3, hexamer, filtered | |||||||||
 Map data Map data | Gaussian filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ / cryoEM / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationRIG-I signaling pathway / granulocyte differentiation / ATP-dependent protein disaggregase activity / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / antiviral innate immune response / mitochondrial intermembrane space / cellular response to heat / ATP hydrolysis activity / mitochondrion / extracellular region ...RIG-I signaling pathway / granulocyte differentiation / ATP-dependent protein disaggregase activity / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / antiviral innate immune response / mitochondrial intermembrane space / cellular response to heat / ATP hydrolysis activity / mitochondrion / extracellular region / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Rizo AN / Cupo RR | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Unique structural features govern the activity of a human mitochondrial AAA+ disaggregase, Skd3. Authors: Ryan R Cupo / Alexandrea N Rizo / Gabriel A Braun / Eric Tse / Edward Chuang / Kushol Gupta / Daniel R Southworth / James Shorter /  Abstract: The AAA+ protein, Skd3 (human CLPB), solubilizes proteins in the mitochondrial intermembrane space, which is critical for human health. Skd3 variants with defective protein-disaggregase activity ...The AAA+ protein, Skd3 (human CLPB), solubilizes proteins in the mitochondrial intermembrane space, which is critical for human health. Skd3 variants with defective protein-disaggregase activity cause severe congenital neutropenia (SCN) and 3-methylglutaconic aciduria type 7 (MGCA7). How Skd3 disaggregates proteins remains poorly understood. Here, we report a high-resolution structure of a Skd3-substrate complex. Skd3 adopts a spiral hexameric arrangement that engages substrate via pore-loop interactions in the nucleotide-binding domain (NBD). Substrate-bound Skd3 hexamers stack head-to-head via unique, adaptable ankyrin-repeat domain (ANK)-mediated interactions to form dodecamers. Deleting the ANK linker region reduces dodecamerization and disaggregase activity. We elucidate apomorphic features of the Skd3 NBD and C-terminal domain that regulate disaggregase activity. We also define how Skd3 subunits collaborate to disaggregate proteins. Importantly, SCN-linked subunits sharply inhibit disaggregase activity, whereas MGCA7-linked subunits do not. These advances illuminate Skd3 structure and mechanism, explain SCN and MGCA7 inheritance patterns, and suggest therapeutic strategies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26122.map.gz emd_26122.map.gz | 140.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26122-v30.xml emd-26122-v30.xml emd-26122.xml emd-26122.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26122.png emd_26122.png | 82 KB | ||
| Filedesc metadata |  emd-26122.cif.gz emd-26122.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26122 http://ftp.pdbj.org/pub/emdb/structures/EMD-26122 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26122 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26122 | HTTPS FTP |
-Validation report
| Summary document |  emd_26122_validation.pdf.gz emd_26122_validation.pdf.gz | 384.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26122_full_validation.pdf.gz emd_26122_full_validation.pdf.gz | 384.5 KB | Display | |
| Data in XML |  emd_26122_validation.xml.gz emd_26122_validation.xml.gz | 7 KB | Display | |
| Data in CIF |  emd_26122_validation.cif.gz emd_26122_validation.cif.gz | 8.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26122 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26122 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26122 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26122 | HTTPS FTP |
-Related structure data
| Related structure data |  7ttsMC  7ttrC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26122.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26122.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Gaussian filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Skd3 bound to FITC-casein
| Entire | Name: Skd3 bound to FITC-casein |
|---|---|
| Components |
|
-Supramolecule #1: Skd3 bound to FITC-casein
| Supramolecule | Name: Skd3 bound to FITC-casein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Macromolecule #1: Caseinolytic peptidase B protein homolog
| Macromolecule | Name: Caseinolytic peptidase B protein homolog / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 66.115047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GGSYSKSPSN KDAALLEAAR ANNMQEVSRL LSEGADVNAK HRLGWTALMV AAINRNNSVV QVLLAAGADP NLGDDFSSVY KTAKEQGIH SLEDGGQDGA SRHITNQWTS ALEFRRWLGL PAGVLITRED DFNNRLNNRA SFKGCTALHY AVLADDYRTV K ELLDGGAN ...String: GGSYSKSPSN KDAALLEAAR ANNMQEVSRL LSEGADVNAK HRLGWTALMV AAINRNNSVV QVLLAAGADP NLGDDFSSVY KTAKEQGIH SLEDGGQDGA SRHITNQWTS ALEFRRWLGL PAGVLITRED DFNNRLNNRA SFKGCTALHY AVLADDYRTV K ELLDGGAN PLQRNEMGHT PLDYAREGEV MKLLRTSEAK YQEKQRKREA EERRRFPLEQ RLKEHIIGQE SAIATVGAAI RR KENGWYD EEHPLVFLFL GSSGIGKTEL AKQTAKYMHK DAKKGFIRLD MSEFQERHEV AKFIGSPPGY VGHEEGGQLT KKL KQCPNA VVLFDEVDKA HPDVLTIMLQ LFDEGRLTDG KGKTIDCKDA IFIMTSNVAS DEIAQHALQL RQEALEMSRN RIAE NLGDV QISDKITISK NFKENVIRPI LKAHFRRDEF LGRINEIVYF LPFCHSELIQ LVNKELNFWA KRAKQRHNIT LLWDR EVAD VLVDGYNVHY GARSIKHEVE RRVVNQLAAA YEQDLLPGGC TLRITVEDSD KQLLKSPELP SPQAEKRLPK LRLEII DKD SKTRRLDIRA PLHPEKVCNT I UniProtKB: Mitochondrial disaggregase |
-Macromolecule #2: Beta-casein
| Macromolecule | Name: Beta-casein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.759648 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)ARELEE LNVP GEIVE SLSSSEESIT RINKKIEKFQ SEEQQQTEDE LQDKIHPFAQ TQSLVYPFPG PIPNSLPQNI PPLTQTPVVV PPFLQ PEVM GVSKVKGAMA ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)ARELEE LNVP GEIVE SLSSSEESIT RINKKIEKFQ SEEQQQTEDE LQDKIHPFAQ TQSLVYPFPG PIPNSLPQNI PPLTQTPVVV PPFLQ PEVM GVSKVKGAMA PKHKEMPFPK YPVEPLTESQ SLTLTDVENL HLPLPLLQSW MHQPHQPLPP TVMFPPQSVL SLSQSK VLP VPQKAVPYPQ RDMPIQAFLL YQEPVLGPVR GPFPIIV UniProtKB: Beta-casein |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 5 / Number of copies: 4 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 68.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7tts: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















