[English] 日本語
 Yorodumi
Yorodumi- EMDB-26072: Coxsackievirus A21 capsid subdomain in complex with mouse polyclo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Coxsackievirus A21 capsid subdomain in complex with mouse polyclonal antibody pAbC-1 | |||||||||
 Map data Map data | Coxsackievirus A21 capsid subdomain in complex with mouse polyclonal antibody pAbC-1 - Main Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | polyclonal antibodies / immune complex / cryoEM / VIRUS-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Coxsackievirus A21 / Coxsackievirus A21 /  | |||||||||
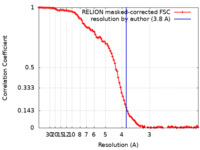
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Antanasijevic A / Ward AB | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: PNAS Nexus / Year: 2022 Journal: PNAS Nexus / Year: 2022Title: High-resolution structural analysis of enterovirus-reactive polyclonal antibodies in complex with whole virions. Authors: Aleksandar Antanasijevic / Autumn J Schulze / Vijay S Reddy / Andrew B Ward /  Abstract: Non-polio enteroviruses (NPEVs) cause serious illnesses in young children and neonates, including aseptic meningitis, encephalitis, and inflammatory muscle disease, among others. While over 100 ...Non-polio enteroviruses (NPEVs) cause serious illnesses in young children and neonates, including aseptic meningitis, encephalitis, and inflammatory muscle disease, among others. While over 100 serotypes have been described to date, vaccine only exists for EV-A71. Efforts toward rationally designed pan-NPEV vaccines would greatly benefit from structural biology methods for rapid and comprehensive evaluation of vaccine candidates and elicited antibody responses. Toward this goal, we introduced a cryo-electron-microscopy-based approach for structural analysis of virus- or vaccine-elicited polyclonal antibodies (pAbs) in complex with whole NPEV virions. We demonstrated the feasibility using coxsackievirus A21 and reconstructed five structurally distinct pAbs bound to the virus. The pAbs targeted two immunodominant epitopes, one overlapping with the receptor binding site. These results demonstrate that our method can be applied to map broad-spectrum polyclonal immune responses against intact virions and define potentially cross-reactive epitopes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26072.map.gz emd_26072.map.gz | 78.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26072-v30.xml emd-26072-v30.xml emd-26072.xml emd-26072.xml | 26.5 KB 26.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26072_fsc.xml emd_26072_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_26072.png emd_26072.png | 121.5 KB | ||
| Masks |  emd_26072_msk_1.map emd_26072_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26072.cif.gz emd-26072.cif.gz | 6.9 KB | ||
| Others |  emd_26072_half_map_1.map.gz emd_26072_half_map_1.map.gz emd_26072_half_map_2.map.gz emd_26072_half_map_2.map.gz | 65.4 MB 65.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26072 http://ftp.pdbj.org/pub/emdb/structures/EMD-26072 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26072 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26072 | HTTPS FTP |
-Validation report
| Summary document |  emd_26072_validation.pdf.gz emd_26072_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26072_full_validation.pdf.gz emd_26072_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_26072_validation.xml.gz emd_26072_validation.xml.gz | 17 KB | Display | |
| Data in CIF |  emd_26072_validation.cif.gz emd_26072_validation.cif.gz | 22.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26072 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26072 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26072 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26072 | HTTPS FTP |
-Related structure data
| Related structure data |  7tquMC  7tqsC  7tqtC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26072.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26072.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Coxsackievirus A21 capsid subdomain in complex with mouse polyclonal antibody pAbC-1 - Main Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
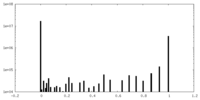
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26072_msk_1.map emd_26072_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
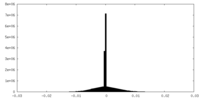
| Density Histograms |
-Half map: Coxsackievirus A21 capsid subdomain in complex with mouse...
| File | emd_26072_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Coxsackievirus A21 capsid subdomain in complex with mouse polyclonal antibody pAbC-1 - Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
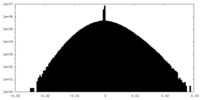
| Density Histograms |
-Half map: Coxsackievirus A21 capsid subdomain in complex with mouse...
| File | emd_26072_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Coxsackievirus A21 capsid subdomain in complex with mouse polyclonal antibody pAbC-1 - Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
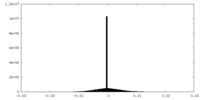
| Density Histograms |
- Sample components
Sample components
-Entire : Immune complex featuring subdomains of coxsackievirus A21 (CV-A21...
| Entire | Name: Immune complex featuring subdomains of coxsackievirus A21 (CV-A21) viral particles in complex with mouse polyclonal antibodies |
|---|---|
| Components |
|
-Supramolecule #1: Immune complex featuring subdomains of coxsackievirus A21 (CV-A21...
| Supramolecule | Name: Immune complex featuring subdomains of coxsackievirus A21 (CV-A21) viral particles in complex with mouse polyclonal antibodies type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 Details: The complexes were assembled by combining purified CV-A21 particles and polyclonal antibodies isolated from mice (as Fab fragments) |
|---|---|
| Source (natural) | Organism:  Coxsackievirus A21 Coxsackievirus A21 |
-Macromolecule #1: pAbC-1 heavy chain
| Macromolecule | Name: pAbC-1 heavy chain / type: protein_or_peptide / ID: 1 / Details: Heavy Chain of the polyclonal antibody pAbC-1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.549763 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #2: pAbC-1 light chain
| Macromolecule | Name: pAbC-1 light chain / type: protein_or_peptide / ID: 2 / Details: Light Chain of the polyclonal antibody pAbC-1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.698714 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 3 / Details: VP1 protein of the coxsackievirus A21 capsid / Number of copies: 3 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Coxsackievirus A21 Coxsackievirus A21 |
| Molecular weight | Theoretical: 33.234238 KDa |
| Sequence | String: GIEDLIDTAI KNALRVSQPP STQSTEATSG VNSQEVPALT AVETGASGQA IPSDVVETRH VVNYKTRSES CLESFFGRAA CVTILSLTN SSKSGEEKKH FNIWNITYTD TVQLRRKLEF FTYSRFDLEM TFVFTENYPS TASGEVRNQV YQIMYIPPGA P RPSSWDDY ...String: GIEDLIDTAI KNALRVSQPP STQSTEATSG VNSQEVPALT AVETGASGQA IPSDVVETRH VVNYKTRSES CLESFFGRAA CVTILSLTN SSKSGEEKKH FNIWNITYTD TVQLRRKLEF FTYSRFDLEM TFVFTENYPS TASGEVRNQV YQIMYIPPGA P RPSSWDDY TWQSSSNPSI FYMYGNAPPR MSIPYVGIAN AYSHFYDGFA RVPLEGENTD AGDTFYGLVS INDFGVLAVR AV NRSNPHT IHTSVRVYMK PKHIRCWCPR PPRAVLYRGE GVDMISSAIL PLAKVDSITT F UniProtKB: Genome polyprotein |
-Macromolecule #4: VP2
| Macromolecule | Name: VP2 / type: protein_or_peptide / ID: 4 / Details: VP2 protein of the coxsackievirus A21 capsid / Number of copies: 3 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Coxsackievirus A21 Coxsackievirus A21 |
| Molecular weight | Theoretical: 29.918537 KDa |
| Sequence | String: SPNVEACGYS DRVRQITLGN STITTQEAAN AIVAYGEWPT YINDSEANPV DAPTEPDVSS NRFYTLESVS WKTTSRGWWW KLPDCLKDM GMFGQNMYYH YLGRSGYTIH VQCNASKFHQ GALGVFLIPE FVMACNTESK TSYVSYINAN PGERGGEFTN T YNPSNTDA ...String: SPNVEACGYS DRVRQITLGN STITTQEAAN AIVAYGEWPT YINDSEANPV DAPTEPDVSS NRFYTLESVS WKTTSRGWWW KLPDCLKDM GMFGQNMYYH YLGRSGYTIH VQCNASKFHQ GALGVFLIPE FVMACNTESK TSYVSYINAN PGERGGEFTN T YNPSNTDA SEGRKFAALD YLLGSGVLAG NAFVYPHQII NLRTNNSATI VVPYVNSLVI DCMAKHNNWG IVILPLAPLA FA ATSSPQV PITVTIAPMC TEFNGLRNIT VPVHQ UniProtKB: Genome polyprotein |
-Macromolecule #5: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 5 / Details: VP3 protein of the coxsackievirus A21 capsid / Number of copies: 3 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Coxsackievirus A21 Coxsackievirus A21 |
| Molecular weight | Theoretical: 26.596703 KDa |
| Sequence | String: GLPTMNTPGS NQFLTSDDFQ SPCALPNFDV TPPIHIPGEV KNMMELAEID TLIPMNAVDG KVNTMEMYQI PLNDNLSKAP IFCLSLSPA SDKRLSHTML GEILNYYTHW TGSIRFTFLF CGSMMATGKL LLSYSPPGAK PPTNRKDAML GTHIIWDLGL Q SSCSMVAP ...String: GLPTMNTPGS NQFLTSDDFQ SPCALPNFDV TPPIHIPGEV KNMMELAEID TLIPMNAVDG KVNTMEMYQI PLNDNLSKAP IFCLSLSPA SDKRLSHTML GEILNYYTHW TGSIRFTFLF CGSMMATGKL LLSYSPPGAK PPTNRKDAML GTHIIWDLGL Q SSCSMVAP WISNTVYRRC ARDDFTEGGF ITCFYQTRIV VPASTPTSMF MLGFVSACPD FSVRLLRDTP HISQSKLIGR TQ UniProtKB: Genome polyprotein |
-Macromolecule #6: VP4
| Macromolecule | Name: VP4 / type: protein_or_peptide / ID: 6 / Details: VP4 protein of the coxsackievirus A21 capsid / Number of copies: 3 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Coxsackievirus A21 Coxsackievirus A21 |
| Molecular weight | Theoretical: 7.442118 KDa |
| Sequence | String: MGAQVSTQKT GAHENQNVAA NGSTINYTTI NYYKDSASNS ATRQDLSQDP SKFTEPVKDL MLKTAPALN UniProtKB: Genome polyprotein |
-Macromolecule #7: MYRISTIC ACID
| Macromolecule | Name: MYRISTIC ACID / type: ligand / ID: 7 / Number of copies: 3 / Formula: MYR |
|---|---|
| Molecular weight | Theoretical: 228.371 Da |
| Chemical component information |  ChemComp-MYR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: TBS, 0.2um filtered | |||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 10 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: 3-7s blot time. | |||||||||
| Details | The complex was generated by incubation of mouse polyclonal antibodies with recombinantly expressed CV-A21 virions and subsequent SEC purification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 3862 / Average exposure time: 9.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)