[English] 日本語
 Yorodumi
Yorodumi- EMDB-25894: Single-molecule 3D density map of HIV cellular entry by liquid-ph... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-molecule 3D density map of HIV cellular entry by liquid-phase electron tomography (particle #3) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV cellular entry / CELL INVASION | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus Human immunodeficiency virus | ||||||||||||
| Method | electron tomography / Resolution: 65.0 Å | ||||||||||||
 Authors Authors | Kong L / Ren G | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Facile hermetic TEM grid preparation for molecular imaging of hydrated biological samples at room temperature. Authors: Lingli Kong / Jianfang Liu / Meng Zhang / Zhuoyang Lu / Han Xue / Amy Ren / Jiankang Liu / Jinping Li / Wai Li Ling / Gang Ren /    Abstract: Although structures of vitrified supramolecular complexes have been determined at near-atomic resolution, elucidating in situ molecular structure in living cells remains a challenge. Here, we report ...Although structures of vitrified supramolecular complexes have been determined at near-atomic resolution, elucidating in situ molecular structure in living cells remains a challenge. Here, we report a straightforward liquid cell technique, originally developed for real-time visualization of dynamics at a liquid-gas interface using transmission electron microscopy, to image wet biological samples. Due to the scattering effects from the liquid phase, the micrographs display an amplitude contrast comparable to that observed in negatively stained samples. We succeed in resolving subunits within the protein complex GroEL imaged in a buffer solution at room temperature. Additionally, we capture various stages of virus cell entry, a process for which only sparse structural data exists due to their transient nature. To scrutinize the morphological details further, we used individual particle electron tomography for 3D reconstruction of each virus. These findings showcase this approach potential as an efficient, cost-effective complement to other microscopy technique in addressing biological questions at the molecular level. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25894.map.gz emd_25894.map.gz | 7.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25894-v30.xml emd-25894-v30.xml emd-25894.xml emd-25894.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
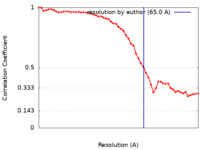
| FSC (resolution estimation) |  emd_25894_fsc.xml emd_25894_fsc.xml | 5.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_25894.png emd_25894.png | 104.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25894 http://ftp.pdbj.org/pub/emdb/structures/EMD-25894 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25894 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25894 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25894.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25894.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 21.44 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : A HIV totally enter cell membrane
| Entire | Name: A HIV totally enter cell membrane |
|---|---|
| Components |
|
-Supramolecule #1: A HIV totally enter cell membrane
| Supramolecule | Name: A HIV totally enter cell membrane / type: organelle_or_cellular_component / ID: 1 / Parent: 0 Details: In the penetrating stage of viral entry, in which a virus half embeds into the cell membrane. |
|---|
-Supramolecule #3: cell membrane
| Supramolecule | Name: cell membrane / type: organelle_or_cellular_component / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Human immunodeficiency virus
| Supramolecule | Name: Human immunodeficiency virus / type: virus / ID: 2 / Parent: 1 / NCBI-ID: 12721 / Sci species name: Human immunodeficiency virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
 Processing Processing | electron tomography |
|---|---|
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: HeLa cells were grown in Gibco minimum essential media (MEM, containing 10 percent FBS) at 37 degree with 5 percent CO2, and virus-infected cells were prepared by incubating HeLa cells with ...Details: HeLa cells were grown in Gibco minimum essential media (MEM, containing 10 percent FBS) at 37 degree with 5 percent CO2, and virus-infected cells were prepared by incubating HeLa cells with virus at a ratio of 50 to 1 for 12 hours. | ||||||
|---|---|---|---|---|---|---|---|
| Sugar embedding | Material: sanwiched by two formvar films Details: sample solution (native liquid state, label-free, without any staining) was deposited and then sandwiched between two layers of 300 mesh TEM Formvar-coated copper grids at room temperature. | ||||||
| Grid | Model: Homemade / Material: COPPER / Support film - Material: FORMVAR / Support film - topology: CONTINUOUS | ||||||
| Details | HeLa cells were grown in Gibco minimum essential media and virus-infected cells were prepared by incubating HeLa cells with virus at a ratio of 50:1 for 12 hours. | ||||||
| Cryo protectant | Liquid-phase | ||||||
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | ZEISS LIBRA120PLUS |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 97 / Average electron dose: 1.0 e/Å2 Details: The total illumination electron doses were ~30 electron per angstrom square. |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated defocus max: 11.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.4 mm / Nominal defocus max: 11.0 µm / Nominal defocus min: 3.0 µm |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)