+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 201phi2-1 Chimallin C1 localized reconstruction | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | phage / viral protein / STRUCTURAL PROTEIN | |||||||||||||||
| Function / homology | host cell cytoplasm / Chimallin Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Pseudomonas phage 201phi2-1 (virus) Pseudomonas phage 201phi2-1 (virus) | |||||||||||||||
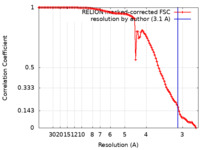
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||
 Authors Authors | Laughlin TG / Deep A / Prichard AM / Seitz C / Gu Y / Enustun E / Suslov S / Khanna K / Birkholz EA / Amaro RE ...Laughlin TG / Deep A / Prichard AM / Seitz C / Gu Y / Enustun E / Suslov S / Khanna K / Birkholz EA / Amaro RE / Pogliano J / Corbett KD / Villa E | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Architecture and self-assembly of the jumbo bacteriophage nuclear shell. Authors: Thomas G Laughlin / Amar Deep / Amy M Prichard / Christian Seitz / Yajie Gu / Eray Enustun / Sergey Suslov / Kanika Khanna / Erica A Birkholz / Emily Armbruster / J Andrew McCammon / Rommie ...Authors: Thomas G Laughlin / Amar Deep / Amy M Prichard / Christian Seitz / Yajie Gu / Eray Enustun / Sergey Suslov / Kanika Khanna / Erica A Birkholz / Emily Armbruster / J Andrew McCammon / Rommie E Amaro / Joe Pogliano / Kevin D Corbett / Elizabeth Villa /  Abstract: Bacteria encode myriad defences that target the genomes of infecting bacteriophage, including restriction-modification and CRISPR-Cas systems. In response, one family of large bacteriophages uses a ...Bacteria encode myriad defences that target the genomes of infecting bacteriophage, including restriction-modification and CRISPR-Cas systems. In response, one family of large bacteriophages uses a nucleus-like compartment to protect its replicating genomes by excluding host defence factors. However, the principal composition and structure of this compartment remain unknown. Here we find that the bacteriophage nuclear shell assembles primarily from one protein, which we name chimallin (ChmA). Combining cryo-electron tomography of nuclear shells in bacteriophage-infected cells and cryo-electron microscopy of a minimal chimallin compartment in vitro, we show that chimallin self-assembles as a flexible sheet into closed micrometre-scale compartments. The architecture and assembly dynamics of the chimallin shell suggest mechanisms for its nucleation and growth, and its role as a scaffold for phage-encoded factors mediating macromolecular transport, cytoskeletal interactions, and viral maturation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25392.map.gz emd_25392.map.gz | 52.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25392-v30.xml emd-25392-v30.xml emd-25392.xml emd-25392.xml | 24.2 KB 24.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25392_fsc.xml emd_25392_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_25392.png emd_25392.png | 92.6 KB | ||
| Masks |  emd_25392_msk_1.map emd_25392_msk_1.map | 105.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25392.cif.gz emd-25392.cif.gz | 6.5 KB | ||
| Others |  emd_25392_additional_1.map.gz emd_25392_additional_1.map.gz emd_25392_half_map_1.map.gz emd_25392_half_map_1.map.gz emd_25392_half_map_2.map.gz emd_25392_half_map_2.map.gz | 1.4 MB 97.5 MB 97.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25392 http://ftp.pdbj.org/pub/emdb/structures/EMD-25392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25392 | HTTPS FTP |
-Validation report
| Summary document |  emd_25392_validation.pdf.gz emd_25392_validation.pdf.gz | 705.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25392_full_validation.pdf.gz emd_25392_full_validation.pdf.gz | 704.6 KB | Display | |
| Data in XML |  emd_25392_validation.xml.gz emd_25392_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_25392_validation.cif.gz emd_25392_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25392 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25392 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25392 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25392 | HTTPS FTP |
-Related structure data
| Related structure data |  7sqsMC  7sqqC  7sqrC  7sqtC  7squC  7sqvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-10862 (Title: Cryo-EM of recombinant 201phi2-1 chimallin / Data size: 679.9 EMPIAR-10862 (Title: Cryo-EM of recombinant 201phi2-1 chimallin / Data size: 679.9 Data #1: Unaligned frames as unormalized LZW-TIFF [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25392.map.gz / Format: CCP4 / Size: 105.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25392.map.gz / Format: CCP4 / Size: 105.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
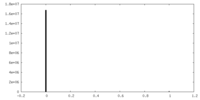
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35265 Å | ||||||||||||||||||||||||||||||||||||
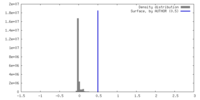
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25392_msk_1.map emd_25392_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
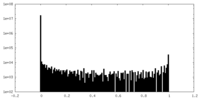
| Density Histograms |
-Additional map: sharpened, reboxed
| File | emd_25392_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened, reboxed | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25392_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_25392_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 201phi2-1 chimallin C1 localized reconstruction
| Entire | Name: 201phi2-1 chimallin C1 localized reconstruction |
|---|---|
| Components |
|
-Supramolecule #1: 201phi2-1 chimallin C1 localized reconstruction
| Supramolecule | Name: 201phi2-1 chimallin C1 localized reconstruction / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage 201phi2-1 (virus) Pseudomonas phage 201phi2-1 (virus) |
-Macromolecule #1: Chimallin
| Macromolecule | Name: Chimallin / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage 201phi2-1 (virus) Pseudomonas phage 201phi2-1 (virus) |
| Molecular weight | Theoretical: 69.784523 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMIRDTAT NTTQTQAAPQ QAPAQQFTQA PQEKPMQSTQ SQPTPSYAGT GGINSQFTRS GNVQGGDARA SEALTVFTRL KEQAVAQQD LADDFSILRF DRDQHQVGWS SLVIAKQISL NGQPVIAVRP LILPNNSIEL PKRKTNIVNG MQTDVIESDI D VGTVFSAQ ...String: SNAMIRDTAT NTTQTQAAPQ QAPAQQFTQA PQEKPMQSTQ SQPTPSYAGT GGINSQFTRS GNVQGGDARA SEALTVFTRL KEQAVAQQD LADDFSILRF DRDQHQVGWS SLVIAKQISL NGQPVIAVRP LILPNNSIEL PKRKTNIVNG MQTDVIESDI D VGTVFSAQ YFNRLSTYVQ NTLGKPGAKV VLAGPFPIPA DLVLKDSELQ LRNLLIKSVN ACDDILALHS GERPFTIAGL KG QQGETLA AKVDIRTQPL HDTVGNPIRA DIVVTTQRVR RNGQQENEFY ETDVKLNQVA MFTNLERTPQ AQAQTLFPNQ QQV ATPAPW VASVVITDVR NADGIQANTP EMYWFALSNA FRSTHGHAWA RPFLPMTGVA KDMKDIGALG WMSALRNRID TKAA NFDDA QFGQLMLSQV QPNPVFQIDL NRMGETAQMD SLQLDAAGGP NAQKAAATII RQINNLGGGG FERFFDHTTQ PILER TGQV IDLGNWFDGD EKRDRRDLDN LAALNAAEGN ENEFWGFYGA QLNPNLHPDL RNRQSRNYDR QYLGSTVTYT GKAERC TYN AKFIEALDRY LAEAGLQITM DNTSVLNSGQ RFMGNSVIGN NMVSGQAQVH SAYAGTQGFN TQYQTGPSSF Y UniProtKB: Chimallin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.019 kPa / Details: 20 mA |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 4192 / Average electron dose: 42.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 108 |
|---|---|
| Output model |  PDB-7sqs: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)