[English] 日本語
 Yorodumi
Yorodumi- EMDB-25228: H. neapolitanus carboxysomal rubisco/CsoSCA-peptide (1-50)complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | H. neapolitanus carboxysomal rubisco/CsoSCA-peptide (1-50)complex | |||||||||
 Map data Map data | upsampled, density-modified map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | rubisco / lyase / TIM-barrel / complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationcarboxysome / ribulose-bisphosphate carboxylase / ribulose-bisphosphate carboxylase activity / reductive pentose-phosphate cycle / carbon fixation / carbonic anhydrase / carbonate dehydratase activity / monooxygenase activity / magnesium ion binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Halothiobacillus neapolitanus (strain ATCC 23641 / c2) (bacteria) Halothiobacillus neapolitanus (strain ATCC 23641 / c2) (bacteria) | |||||||||
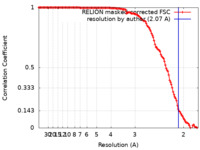
| Method | single particle reconstruction / cryo EM / Resolution: 2.07 Å | |||||||||
 Authors Authors | Blikstad C / Dugan E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Identification of a carbonic anhydrase-Rubisco complex within the alpha-carboxysome. Authors: Cecilia Blikstad / Eli J Dugan / Thomas G Laughlin / Julia B Turnšek / Mira D Liu / Sophie R Shoemaker / Nikoleta Vogiatzi / Jonathan P Remis / David F Savage /   Abstract: Carboxysomes are proteinaceous organelles that encapsulate key enzymes of CO fixation-Rubisco and carbonic anhydrase-and are the centerpiece of the bacterial CO concentrating mechanism (CCM). In the ...Carboxysomes are proteinaceous organelles that encapsulate key enzymes of CO fixation-Rubisco and carbonic anhydrase-and are the centerpiece of the bacterial CO concentrating mechanism (CCM). In the CCM, actively accumulated cytosolic bicarbonate diffuses into the carboxysome and is converted to CO by carbonic anhydrase, producing a high CO concentration near Rubisco and ensuring efficient carboxylation. Self-assembly of the α-carboxysome is orchestrated by the intrinsically disordered scaffolding protein, CsoS2, which interacts with both Rubisco and carboxysomal shell proteins, but it is unknown how the carbonic anhydrase, CsoSCA, is incorporated into the α-carboxysome. Here, we present the structural basis of carbonic anhydrase encapsulation into α-carboxysomes from . We find that CsoSCA interacts directly with Rubisco via an intrinsically disordered N-terminal domain. A 1.98 Å single-particle cryoelectron microscopy structure of Rubisco in complex with this peptide reveals that CsoSCA binding is predominantly mediated by a network of hydrogen bonds. CsoSCA's binding site overlaps with that of CsoS2, but the two proteins utilize substantially different motifs and modes of binding, revealing a plasticity of the Rubisco binding site. Our results advance the understanding of carboxysome biogenesis and highlight the importance of Rubisco, not only as an enzyme but also as a central hub for mediating assembly through protein interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25228.map.gz emd_25228.map.gz | 50.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25228-v30.xml emd-25228-v30.xml emd-25228.xml emd-25228.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25228_fsc.xml emd_25228_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_25228.png emd_25228.png | 256.1 KB | ||
| Masks |  emd_25228_msk_1.map emd_25228_msk_1.map | 193.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25228.cif.gz emd-25228.cif.gz | 6.7 KB | ||
| Others |  emd_25228_additional_1.map.gz emd_25228_additional_1.map.gz emd_25228_half_map_1.map.gz emd_25228_half_map_1.map.gz emd_25228_half_map_2.map.gz emd_25228_half_map_2.map.gz | 151.2 MB 147.1 MB 147.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25228 http://ftp.pdbj.org/pub/emdb/structures/EMD-25228 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25228 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25228 | HTTPS FTP |
-Related structure data
| Related structure data |  7snvMC  7smkC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25228.map.gz / Format: CCP4 / Size: 54.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25228.map.gz / Format: CCP4 / Size: 54.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | upsampled, density-modified map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.60059 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25228_msk_1.map emd_25228_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
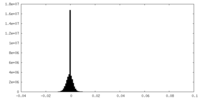
| Density Histograms |
-Additional map: unmodified full map
| File | emd_25228_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unmodified full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
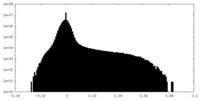
| Density Histograms |
-Half map: Carboxysomal rubisco/CsoSCA-peptide (1-50)complex
| File | emd_25228_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Carboxysomal rubisco/CsoSCA-peptide (1-50)complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
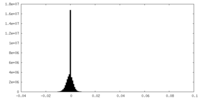
| Density Histograms |
-Half map: Carboxysomal rubisco/CsoSCA-peptide (1-50)complex
| File | emd_25228_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Carboxysomal rubisco/CsoSCA-peptide (1-50)complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
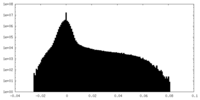
| Density Histograms |
- Sample components
Sample components
-Entire : H. neapolitanus carboxysomal rubisco/CsoSCA-peptide (1-50)complex
| Entire | Name: H. neapolitanus carboxysomal rubisco/CsoSCA-peptide (1-50)complex |
|---|---|
| Components |
|
-Supramolecule #1: H. neapolitanus carboxysomal rubisco/CsoSCA-peptide (1-50)complex
| Supramolecule | Name: H. neapolitanus carboxysomal rubisco/CsoSCA-peptide (1-50)complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 533 KDa |
-Supramolecule #2: Ribulose bisphosphate carboxylase large chain (E.C.4.1.1.39), Rib...
| Supramolecule | Name: Ribulose bisphosphate carboxylase large chain (E.C.4.1.1.39), Ribulose bisphosphate carboxylase small chain (E.C.4.1.1.39) type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Halothiobacillus neapolitanus (strain ATCC 23641 / c2) (bacteria) Halothiobacillus neapolitanus (strain ATCC 23641 / c2) (bacteria) |
-Supramolecule #3: Carboxysome shell carbonic anhydrase (E.C.4.2.1.1)
| Supramolecule | Name: Carboxysome shell carbonic anhydrase (E.C.4.2.1.1) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|
-Macromolecule #1: Ribulose bisphosphate carboxylase large chain
| Macromolecule | Name: Ribulose bisphosphate carboxylase large chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: ribulose-bisphosphate carboxylase |
|---|---|
| Source (natural) | Organism:  Halothiobacillus neapolitanus (strain ATCC 23641 / c2) (bacteria) Halothiobacillus neapolitanus (strain ATCC 23641 / c2) (bacteria)Strain: ATCC 23641 / c2 |
| Molecular weight | Theoretical: 53.831723 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAVKKYSAG VKEYRQTYWM PEYTPLDSDI LACFKITPQP GVDREEAAAA VAAESSTGTW TTVWTDLLTD MDYYKGRAYR IEDVPGDDA AFYAFIAYPI DLFEEGSVVN VFTSLVGNVF GFKAVRGLRL EDVRFPLAYV KTCGGPPHGI QVERDKMNKY G RPLLGCTI ...String: MSAVKKYSAG VKEYRQTYWM PEYTPLDSDI LACFKITPQP GVDREEAAAA VAAESSTGTW TTVWTDLLTD MDYYKGRAYR IEDVPGDDA AFYAFIAYPI DLFEEGSVVN VFTSLVGNVF GFKAVRGLRL EDVRFPLAYV KTCGGPPHGI QVERDKMNKY G RPLLGCTI KPKLGLSAKN YGRAVYECLR GGLDFTKDDE NINSQPFMRW RDRFLFVQDA TETAEAQTGE RKGHYLNVTA PT PEEMYKR AEFAKEIGAP IIMHDYITGG FTANTGLAKW CQDNGVLLHI HRAMHAVIDR NPNHGIHFRV LTKILRLSGG DHL HTGTVV GKLEGDRAST LGWIDLLRES FIPEDRSRGI FFDQDWGSMP GVFAVASGGI HVWHMPALVN IFGDDSVLQF GGGT LGHPW GNAAGAAANR VALEACVEAR NQGRDIEKEG KEILTAAAQH SPELKIAMET WKEIKFEFDT VDKLDTQNRW SHPQF EK UniProtKB: Ribulose bisphosphate carboxylase large chain |
-Macromolecule #2: Ribulose bisphosphate carboxylase small chain
| Macromolecule | Name: Ribulose bisphosphate carboxylase small chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: ribulose-bisphosphate carboxylase |
|---|---|
| Source (natural) | Organism:  Halothiobacillus neapolitanus (strain ATCC 23641 / c2) (bacteria) Halothiobacillus neapolitanus (strain ATCC 23641 / c2) (bacteria)Strain: ATCC 23641 / c2 |
| Molecular weight | Theoretical: 12.866575 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAEMQDYKQS LKYETFSYLP PMNAERIRAQ IKYAIAQGWS PGIEHVEVKN SMNQYWYMWK LPFFGEQNVD NVLAEIEACR SAYPTHQVK LVAYDNYAQS LGLAFVVYRG N UniProtKB: Ribulose bisphosphate carboxylase small subunit |
-Macromolecule #3: Carboxysome shell carbonic anhydrase
| Macromolecule | Name: Carboxysome shell carbonic anhydrase / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: carbonic anhydrase |
|---|---|
| Source (natural) | Organism:  Halothiobacillus neapolitanus (strain ATCC 23641 / c2) (bacteria) Halothiobacillus neapolitanus (strain ATCC 23641 / c2) (bacteria) |
| Molecular weight | Theoretical: 5.690505 KDa |
| Sequence | String: MNTRNTRSKQ RAPFGVSSSV KPRLDLIEQA PNPAYDRHPA CITLPERTCR UniProtKB: Carboxysome shell carbonic anhydrase |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 200 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2665 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 3 seconds. | ||||||||||||
| Details | 0.5 uM of rubisco and 0.5 mM of peptide |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 5742 / Average electron dose: 50.0 e/Å2 / Details: 3 x 3 multishot image shift pattern. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 57000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Map-model FSC and geometry |
|---|---|
| Output model |  PDB-7snv: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)