[English] 日本語
 Yorodumi
Yorodumi- EMDB-25184: Ab initio structure of triclinic lysozyme from electron-counted M... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ab initio structure of triclinic lysozyme from electron-counted MicroED data | |||||||||
 Map data Map data | Ab initio normalized structure factor map of P1 lysozyme after fragment placement and density modification from electron-counted MicroED data | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hydrolase | |||||||||
| Function / homology |  Function and homology information Function and homology informationLactose synthesis / Antimicrobial peptides / Neutrophil degranulation / beta-N-acetylglucosaminidase activity / cell wall macromolecule catabolic process / lysozyme / lysozyme activity / defense response to Gram-negative bacterium / killing of cells of another organism / defense response to Gram-positive bacterium ...Lactose synthesis / Antimicrobial peptides / Neutrophil degranulation / beta-N-acetylglucosaminidase activity / cell wall macromolecule catabolic process / lysozyme / lysozyme activity / defense response to Gram-negative bacterium / killing of cells of another organism / defense response to Gram-positive bacterium / defense response to bacterium / endoplasmic reticulum / extracellular space / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | electron crystallography / cryo EM | |||||||||
 Authors Authors | Martynowycz MW / Clabbers MTB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2022 Journal: Nat Methods / Year: 2022Title: Ab initio phasing macromolecular structures using electron-counted MicroED data. Authors: Michael W Martynowycz / Max T B Clabbers / Johan Hattne / Tamir Gonen /  Abstract: Structures of two globular proteins were determined ab initio using microcrystal electron diffraction (MicroED) data that were collected on a direct electron detector in counting mode. Microcrystals ...Structures of two globular proteins were determined ab initio using microcrystal electron diffraction (MicroED) data that were collected on a direct electron detector in counting mode. Microcrystals were identified using a scanning electron microscope (SEM) and thinned with a focused ion beam (FIB) to produce crystalline lamellae of ideal thickness. Continuous-rotation data were collected using an ultra-low exposure rate to enable electron counting in diffraction. For the first sample, triclinic lysozyme extending to a resolution of 0.87 Å, an ideal helical fragment of only three alanine residues provided initial phases. These phases were improved using density modification, allowing the entire atomic structure to be built automatically. A similar approach was successful on a second macromolecular sample, proteinase K, which is much larger and diffracted to a resolution of 1.5 Å. These results demonstrate that macromolecules can be determined to sub-ångström resolution by MicroED and that ab initio phasing can be successfully applied to counting data. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25184.map.gz emd_25184.map.gz | 10.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25184-v30.xml emd-25184-v30.xml emd-25184.xml emd-25184.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25184.png emd_25184.png | 83 KB | ||
| Filedesc metadata |  emd-25184.cif.gz emd-25184.cif.gz | 5.6 KB | ||
| Others |  emd_25184_additional_1.map.gz emd_25184_additional_1.map.gz emd_25184_additional_2.map.gz emd_25184_additional_2.map.gz | 25.6 MB 25.5 MB | ||
| Filedesc structureFactors |  emd_25184_sf.cif.gz emd_25184_sf.cif.gz | 1.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25184 http://ftp.pdbj.org/pub/emdb/structures/EMD-25184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25184 | HTTPS FTP |
-Related structure data
| Related structure data |  7skwMC  7skxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25184.map.gz / Format: CCP4 / Size: 11 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25184.map.gz / Format: CCP4 / Size: 11 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ab initio normalized structure factor map of P1 lysozyme after fragment placement and density modification from electron-counted MicroED data | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X: 0.21136 Å / Y: 0.21333 Å / Z: 0.20631 Å | ||||||||||||||||||||||||||||||||||||
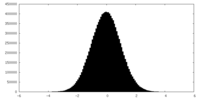
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: 2mFo-dFc map of P1 lysozyme determined ab initio...
| File | emd_25184_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 2mFo-dFc map of P1 lysozyme determined ab initio by electron counted MicroED data | ||||||||||||
| Projections & Slices |
| ||||||||||||
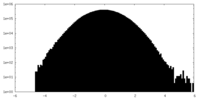
| Density Histograms |
-Additional map: mFo-dFc map of P1 lysozyme determined ab initio...
| File | emd_25184_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mFo-dFc map of P1 lysozyme determined ab initio by electron counted MicroED data | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Lysozyme
| Entire | Name: Lysozyme |
|---|---|
| Components |
|
-Supramolecule #1: Lysozyme
| Supramolecule | Name: Lysozyme / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.4 KDa |
-Macromolecule #1: Lysozyme C
| Macromolecule | Name: Lysozyme C / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: lysozyme |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.33116 KDa |
| Sequence | String: KVFGRCELAA AMKRHGLDNY RGYSLGNWVC AAKFESNFNT QATNRNTDGS TDYGILQINS RWWCNDGRTP GSRNLCNIPC SALLSSDIT ASVNCAKKIV SDGNGMNAWV AWRNRCKGTD VQAWIRGCRL UniProtKB: Lysozyme C |
-Macromolecule #2: NITRATE ION
| Macromolecule | Name: NITRATE ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: NO3 |
|---|---|
| Molecular weight | Theoretical: 62.005 Da |
| Chemical component information |  ChemComp-NO3: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 156 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 4.7 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 10 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: LEICA PLUNGER |
| Details | Milled microcrystals |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 90.0 K |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Number grids imaged: 1 / Number real images: 1 / Number diffraction images: 840 / Average exposure time: 0.5 sec. / Average electron dose: 0.001 e/Å2 Details: 0.15 degrees per second, 0.5 second readout, 30 to -30 degrees |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Cs: 2.7 mm / Camera length: 1202 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN / Tilt angle: -30.0, 30.0 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Binned by 2 |
|---|---|
| Final reconstruction | Resolution method: DIFFRACTION PATTERN/LAYERLINES / Software - Name: REFMAC |
| Merging software list | Software - Name: AIMLESS |
| Crystallography statistics | Number intensities measured: 569407 / Number structure factors: 64974 / Fourier space coverage: 87.58 / R sym: 0.073 / R merge: 0.236 / Overall phase error: 30 / Overall phase residual: 0 / Phase error rejection criteria: None / High resolution: 0.87 Å / Shell - Shell ID: 1 / Shell - High resolution: 0.87 Å / Shell - Low resolution: 0.9 Å / Shell - Number structure factors: 2783 / Shell - Phase residual: 30 / Shell - Fourier space coverage: 37.64 / Shell - Multiplicity: 2.1 |
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL / Overall B value: 12.188 / Target criteria: Maximum likelihood |
|---|---|
| Output model |  PDB-7skw: |
 Movie
Movie Controller
Controller








 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)