[English] 日本語
 Yorodumi
Yorodumi- EMDB-2514: ER membrane-associated ribosome from HeLa cells after treatment w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2514 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ER membrane-associated ribosome from HeLa cells after treatment with non-silencing control siRNA | |||||||||
 Map data Map data | ER membrane-associated ribosome from HeLa cells after treatment with non-silencing control siRNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | endoplasmic reticulum / translocon / ribosome / oligosaccharyltransferase / OST / protein-conducting channel / Sec61 / translocon-associated protein complex / TRAP | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 40.0 Å | |||||||||
 Authors Authors | Pfeffer S / Dudek J / Gogala M / Schorr S / Linxweiler J / Lang S / Becker T / Beckmann R / Zimmermann R / Foerster F | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2014 Journal: Nat Commun / Year: 2014Title: Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Authors: Stefan Pfeffer / Johanna Dudek / Marko Gogala / Stefan Schorr / Johannes Linxweiler / Sven Lang / Thomas Becker / Roland Beckmann / Richard Zimmermann / Friedrich Förster /  Abstract: In mammalian cells, proteins are typically translocated across the endoplasmic reticulum (ER) membrane in a co-translational mode by the ER protein translocon, comprising the protein-conducting ...In mammalian cells, proteins are typically translocated across the endoplasmic reticulum (ER) membrane in a co-translational mode by the ER protein translocon, comprising the protein-conducting channel Sec61 and additional complexes involved in nascent chain processing and translocation. As an integral component of the translocon, the oligosaccharyl-transferase complex (OST) catalyses co-translational N-glycosylation, one of the most common protein modifications in eukaryotic cells. Here we use cryoelectron tomography, cryoelectron microscopy single-particle analysis and small interfering RNA-mediated gene silencing to determine the overall structure, oligomeric state and position of OST in the native ER protein translocon of mammalian cells in unprecedented detail. The observed positioning of OST in close proximity to Sec61 provides a basis for understanding how protein translocation into the ER and glycosylation of nascent proteins are structurally coupled. The overall spatial organization of the native translocon, as determined here, serves as a reliable framework for further hypothesis-driven studies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2514.map.gz emd_2514.map.gz | 28.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2514-v30.xml emd-2514-v30.xml emd-2514.xml emd-2514.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2514.tif emd_2514.tif | 816.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2514 http://ftp.pdbj.org/pub/emdb/structures/EMD-2514 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2514 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2514 | HTTPS FTP |
-Related structure data
| Related structure data |  2515C  2516C  2517C  2518C  2519C  2523C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2514.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2514.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ER membrane-associated ribosome from HeLa cells after treatment with non-silencing control siRNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.88 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
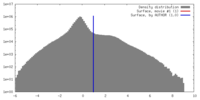
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ER membrane-associated ribosome from HeLa cells after treatment w...
| Entire | Name: ER membrane-associated ribosome from HeLa cells after treatment with non-silencing control siRNA |
|---|---|
| Components |
|
-Supramolecule #1000: ER membrane-associated ribosome from HeLa cells after treatment w...
| Supramolecule | Name: ER membrane-associated ribosome from HeLa cells after treatment with non-silencing control siRNA type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Supramolecule #1: ER membrane-associated 80S ribosome
| Supramolecule | Name: ER membrane-associated 80S ribosome / type: complex / ID: 1 / Recombinant expression: No / Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: HeLa / synonym: Human / Location in cell: Endoplasmic reticulum Homo sapiens (human) / Strain: HeLa / synonym: Human / Location in cell: Endoplasmic reticulum |
-Macromolecule #1: ER protein translocon
| Macromolecule | Name: ER protein translocon / type: protein_or_peptide / ID: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: HeLa / synonym: Human / Location in cell: Endoplasmic reticulum Homo sapiens (human) / Strain: HeLa / synonym: Human / Location in cell: Endoplasmic reticulum |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Grid | Details: Lacey carbon molybdenum grid |
| Vitrification | Cryogen name: ETHANE-PROPANE MIXTURE / Chamber humidity: 70 % / Instrument: FEI VITROBOT MARK IV / Method: Blot 3 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Oct 31, 2012 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON I (4k x 4k) / Average electron dose: 60 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 8.0 µm / Nominal defocus min: 7.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Candidate particles were localized using template matching and cross correlation peaks in areas containing rough ER were visually inspected to identify true positive matches. At the selected coordinates, unbinned subtomograms were reconstructed individually from the weighted back-projections and aligned to a template. |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 40.0 Å / Resolution method: OTHER / Software - Name: av3, tom, PyTom / Number subtomograms used: 295 |
| CTF correction | Details: projection |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)