[English] 日本語
 Yorodumi
Yorodumi- EMDB-25037: Cryo-EM 3D map of the human Exostosin-1 and Exostosin-2 heterodim... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM 3D map of the human Exostosin-1 and Exostosin-2 heterodimer in complex with a 7-sugar oligosaccharide acceptor analog | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | exostosin1 / exostosin2 / glycosyltransferase / heparan sulfate / acceptor / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationglucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase / N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase / hypersensitivity / heart field specification / lymphocyte adhesion to endothelial cell of high endothelial venule / heparan sulfate N-acetylglucosaminyltransferase activity / glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity / N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity / smoothened signaling pathway involved in lung development / developmental growth involved in morphogenesis ...glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase / N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase / hypersensitivity / heart field specification / lymphocyte adhesion to endothelial cell of high endothelial venule / heparan sulfate N-acetylglucosaminyltransferase activity / glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity / N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity / smoothened signaling pathway involved in lung development / developmental growth involved in morphogenesis / sweat gland development / perichondral bone morphogenesis / mesenchymal cell differentiation involved in bone development / response to leukemia inhibitory factor / UDP-N-acetylglucosamine transferase complex / chondrocyte hypertrophy / embryonic skeletal joint development / hematopoietic stem cell migration to bone marrow / fluid transport / tight junction organization / glucuronosyltransferase activity / limb joint morphogenesis / heparin proteoglycan biosynthetic process / Defective EXT2 causes exostoses 2 / Defective EXT1 causes exostoses 1, TRPS2 and CHDS / sebaceous gland development / glomerular basement membrane development / response to heparin / stomach development / glycosaminoglycan biosynthetic process / glandular epithelial cell differentiation / lymphocyte migration into lymphoid organs / HS-GAG biosynthesis / sulfation / chondrocyte proliferation / dendritic cell migration / endochondral bone morphogenesis / dendrite self-avoidance / heparan sulfate proteoglycan biosynthetic process / acetylglucosaminyltransferase activity / endochondral bone growth / podocyte differentiation / hematopoietic stem cell homeostasis / cartilage development involved in endochondral bone morphogenesis / sodium ion homeostasis / basement membrane organization / vocalization behavior / olfactory bulb development / endochondral ossification / vacuole organization / cranial skeletal system development / multicellular organismal-level water homeostasis / endoderm development / leukocyte tethering or rolling / fear response / polysaccharide biosynthetic process / response to light intensity / optic nerve development / protein N-linked glycosylation / regulation of tumor necrosis factor-mediated signaling pathway / collagen fibril organization / ossification involved in bone maturation / cell adhesion mediated by integrin / hair follicle morphogenesis / neural crest cell differentiation / stem cell division / : / heart contraction / antigen processing and presentation / epithelial tube branching involved in lung morphogenesis / glycosyltransferase activity / mesoderm development / motor behavior / mesoderm formation / blood vessel remodeling / fibroblast growth factor receptor signaling pathway / social behavior / cell fate commitment / chondrocyte differentiation / hematopoietic stem cell differentiation / canonical Wnt signaling pathway / bone resorption / BMP signaling pathway / catalytic complex / cellular response to fibroblast growth factor stimulus / gastrulation / axon guidance / ossification / synaptic transmission, glutamatergic / protein catabolic process / wound healing / cellular response to virus / regulation of blood pressure / vasodilation / multicellular organism growth / protein-containing complex assembly / gene expression / protein heterodimerization activity / Golgi membrane / synapse Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
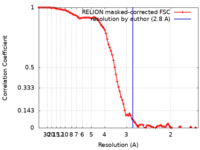
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: Structural basis for heparan sulfate co-polymerase action by the EXT1-2 complex. Authors: Hua Li / Digantkumar Chapla / Robert A Amos / Annapoorani Ramiah / Kelley W Moremen / Huilin Li /  Abstract: Heparan sulfate (HS) proteoglycans are extended (-GlcAβ1,4GlcNAcα1,4-) co-polymers containing decorations of sulfation and epimerization that are linked to cell surface and extracellular matrix ...Heparan sulfate (HS) proteoglycans are extended (-GlcAβ1,4GlcNAcα1,4-) co-polymers containing decorations of sulfation and epimerization that are linked to cell surface and extracellular matrix proteins. In mammals, HS repeat units are extended by an obligate heterocomplex of two exostosin family members, EXT1 and EXT2, where each protein monomer contains distinct GT47 (GT-B fold) and GT64 (GT-A fold) glycosyltransferase domains. In this study, we generated human EXT1-EXT2 (EXT1-2) as a functional heterocomplex and determined its structure in the presence of bound donor and acceptor substrates. Structural data and enzyme activity of catalytic site mutants demonstrate that only two of the four glycosyltransferase domains are major contributors to co-polymer syntheses: the EXT1 GT-B fold β1,4GlcA transferase domain and the EXT2 GT-A fold α1,4GlcNAc transferase domain. The two catalytic sites are over 90 Å apart, indicating that HS is synthesized by a dissociative process that involves a single catalytic site on each monomer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25037.map.gz emd_25037.map.gz | 56.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25037-v30.xml emd-25037-v30.xml emd-25037.xml emd-25037.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25037_fsc.xml emd_25037_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_25037.png emd_25037.png | 162.6 KB | ||
| Masks |  emd_25037_msk_1.map emd_25037_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25037.cif.gz emd-25037.cif.gz | 7 KB | ||
| Others |  emd_25037_additional_1.map.gz emd_25037_additional_1.map.gz emd_25037_half_map_1.map.gz emd_25037_half_map_1.map.gz emd_25037_half_map_2.map.gz emd_25037_half_map_2.map.gz | 6.4 MB 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25037 http://ftp.pdbj.org/pub/emdb/structures/EMD-25037 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25037 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25037 | HTTPS FTP |
-Related structure data
| Related structure data |  7sckMC  7schC  7scjC  7uqxC  7uqyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25037.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25037.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25037_msk_1.map emd_25037_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
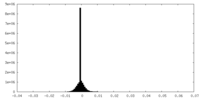
| Density Histograms |
-Additional map: #1
| File | emd_25037_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
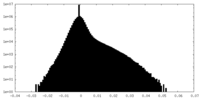
| Density Histograms |
-Half map: #2
| File | emd_25037_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
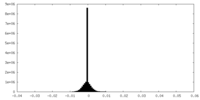
| Density Histograms |
-Half map: #1
| File | emd_25037_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
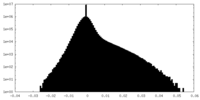
| Density Histograms |
- Sample components
Sample components
-Entire : human EXT1/2
| Entire | Name: human EXT1/2 |
|---|---|
| Components |
|
-Supramolecule #1: human EXT1/2
| Supramolecule | Name: human EXT1/2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Exostosin-1
| Macromolecule | Name: Exostosin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 83.404023 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GFRASRSHSR REEHSGRNGL HHPSPDHFWP RFPDALRPFV PWDQLENEDS SVHISPRQKR DANSSIYKGK KCRMESCFDF TLCKKNGFK VYVYPQQKGE KIAESYQNIL AAIEGSRFYT SDPSQACLFV LSLDTLDRDQ LSPQYVHNLR SKVQSLHLWN N GRNHLIFN ...String: GFRASRSHSR REEHSGRNGL HHPSPDHFWP RFPDALRPFV PWDQLENEDS SVHISPRQKR DANSSIYKGK KCRMESCFDF TLCKKNGFK VYVYPQQKGE KIAESYQNIL AAIEGSRFYT SDPSQACLFV LSLDTLDRDQ LSPQYVHNLR SKVQSLHLWN N GRNHLIFN LYSGTWPDYT EDVGFDIGQA MLAKASISTE NFRPNFDVSI PLFSKDHPRT GGERGFLKFN TIPPLRKYML VF KGKRYLT GIGSDTRNAL YHVHNGEDVV LLTTCKHGKD WQKHKDSRCD RDNTEYEKYD YREMLHNATF CLVPRGRRLG SFR FLEALQ AACVPVMLSN GWELPFSEVI NWNQAAVIGD ERLLLQIPST IRSIHQDKIL ALRQQTQFLW EAYFSSVEKI VLTT LEIIQ DRIFKHISRN SLIWNKHPGG LFVLPQYSSY LGDFPYYYAN LGLKPPSKFT AVIHAVTPLV SQSQPVLKLL VAAAK SQYC AQIIVLWNCD KPLPAKHRWP ATAVPVVVIE GESKVMSSRF LPYDNIITDA VLSLDEDTVL STTEVDFAFT VWQSFP ERI VGYPARSHFW DNSKERWGYT SKWTNDYSMV LTGAAIYHKY YHYLYSHYLP ASLKNMVDQL ANCEDILMNF LVSAVTK LP PIKVTQKKQY KETMMGQTSR ASRWADPDHF AQRQSCMNTF ASWFGYMPLI HSQMRLDPVL FKDQVSILRK KYRDIERL UniProtKB: Exostosin-1 |
-Macromolecule #2: Exostosin-2
| Macromolecule | Name: Exostosin-2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 77.238031 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GWPHSIESSN DWNVEKRSIR DVPVVRLPAD SPIPERGDLS CRMHTCFDVY RCGFNPKNKI KVYIYALKKY VDDFGVSVSN TISREYNEL LMAISDSDYY TDDINRACLF VPSIDVLNQN TLRIKETAQA MAQLSRWDRG TNHLLFNMLP GGPPDYNTAL D VPRDRALL ...String: GWPHSIESSN DWNVEKRSIR DVPVVRLPAD SPIPERGDLS CRMHTCFDVY RCGFNPKNKI KVYIYALKKY VDDFGVSVSN TISREYNEL LMAISDSDYY TDDINRACLF VPSIDVLNQN TLRIKETAQA MAQLSRWDRG TNHLLFNMLP GGPPDYNTAL D VPRDRALL AGGGFSTWTY RQGYDVSIPV YSPLSAEVDL PEKGPGPRQY FLLSSQVGLH PEYREDLEAL QVKHGESVLV LD KCTNLSE GVLSVRKRCH KHQVFDYPQV LQEATFCVVL RGARLGQAVL SDVLQAGCVP VVIADSYILP FSEVLDWKRA SVV VPEEKM SDVYSILQSI PQRQIEEMQR QARWFWEAYF QSIKAIALAT LQIINDRIYP YAAISYEEWN DPPAVKWGSV SNPL FLPLI PPQSQGFTAI VLTYDRVESL FRVITEVSKV PSLSKLLVVW NNQNKNPPED SLWPKIRVPL KVVRTAENKL SNRFF PYDE IETEAVLAID DDIIMLTSDE LQFGYEVWRE FPDRLVGYPG RLHLWDHEMN KWKYESEWTN EVSMVLTGAA FYHKYF NYL YTYKMPGDIK NWVDAHMNCE DIAMNFLVAN VTGKAVIKVT PRKKFKCPEC TAIDGLSLDQ THMVERSECI NKFASVF GT MPLKVVEHRA DPVLYKDDFP EKLKSFPNIG SL UniProtKB: Exostosin-2 |
-Macromolecule #5: URIDINE-5'-DIPHOSPHATE
| Macromolecule | Name: URIDINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: UDP |
|---|---|
| Molecular weight | Theoretical: 404.161 Da |
| Chemical component information |  ChemComp-UDP: |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| |||||||||
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 299 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 193.0 K / Max: 193.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.05 mrad |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Number grids imaged: 1 / #0 - Number real images: 5108 / #0 - Average exposure time: 1.5 sec. / #0 - Average electron dose: 68.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Digitization - Dimensions - Width: 5760 pixel / #1 - Digitization - Dimensions - Height: 4092 pixel / #1 - Number grids imaged: 1 / #1 - Number real images: 7609 / #1 - Average exposure time: 1.5 sec. / #1 - Average electron dose: 72.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7sck: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)