+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24506 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of US27-Gi-scFv16 in CL-state | |||||||||
 Map data Map data | Full local anisotropic sharpen map (Phenix). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Viral GPCR / HCMV / cytomegalovirus / G protein complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationC-C chemokine receptor activity / C-C chemokine binding / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding ...C-C chemokine receptor activity / C-C chemokine binding / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / cell chemotaxis / Regulation of insulin secretion / calcium-mediated signaling / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / virion component / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / GDP binding / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / immune response / ciliary basal body / G protein-coupled receptor signaling pathway / lysosomal membrane / cell division / GTPase activity / synapse / centrosome / GTP binding / protein-containing complex binding / nucleolus / host cell plasma membrane / magnesium ion binding / Golgi apparatus / signal transduction / extracellular exosome / nucleoplasm / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /    Human betaherpesvirus 5 / Human betaherpesvirus 5 /   Human cytomegalovirus Human cytomegalovirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Tsutsumi N / Jude KM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Atypical structural snapshots of human cytomegalovirus GPCR interactions with host G proteins Authors: Tsutsumi N / Maeda S / Qu Q / Voegele M / Jude KM / Suomivuori CM / Panova O / Waghray D / Kato HE / Velasco A / Dror RO / Skiniotis G / Kobilka BK / Garcia KC | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24506.map.gz emd_24506.map.gz | 96.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24506-v30.xml emd-24506-v30.xml emd-24506.xml emd-24506.xml | 29.9 KB 29.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24506.png emd_24506.png | 111.9 KB | ||
| Filedesc metadata |  emd-24506.cif.gz emd-24506.cif.gz | 7.7 KB | ||
| Others |  emd_24506_additional_1.map.gz emd_24506_additional_1.map.gz emd_24506_additional_2.map.gz emd_24506_additional_2.map.gz emd_24506_half_map_1.map.gz emd_24506_half_map_1.map.gz emd_24506_half_map_2.map.gz emd_24506_half_map_2.map.gz | 53.2 MB 51.9 MB 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24506 http://ftp.pdbj.org/pub/emdb/structures/EMD-24506 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24506 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24506 | HTTPS FTP |
-Validation report
| Summary document |  emd_24506_validation.pdf.gz emd_24506_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24506_full_validation.pdf.gz emd_24506_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_24506_validation.xml.gz emd_24506_validation.xml.gz | 13.5 KB | Display | |
| Data in CIF |  emd_24506_validation.cif.gz emd_24506_validation.cif.gz | 16 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24506 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24506 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24506 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24506 | HTTPS FTP |
-Related structure data
| Related structure data |  7rkxMC  7rkfC  7rkmC  7rknC  7rkyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24506.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24506.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full local anisotropic sharpen map (Phenix). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8677 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
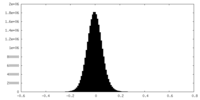
-Supplemental data
-Additional map: Full uniform sharpen map used for refinement.
| File | emd_24506_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full uniform sharpen map used for refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
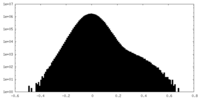
-Additional map: Full unsharpen map.
| File | emd_24506_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full unsharpen map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
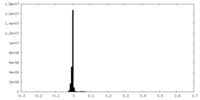
-Half map: Half map B.
| File | emd_24506_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A.
| File | emd_24506_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : US27-Gi-scFv16 complex
| Entire | Name: US27-Gi-scFv16 complex |
|---|---|
| Components |
|
-Supramolecule #1: US27-Gi-scFv16 complex
| Supramolecule | Name: US27-Gi-scFv16 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Gi heterotrimer
| Supramolecule | Name: Gi heterotrimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Location in cell: Membrane Homo sapiens (human) / Location in cell: Membrane |
| Molecular weight | Theoretical: 90 KDa |
-Supramolecule #3: scFv16
| Supramolecule | Name: scFv16 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30 KDa |
-Supramolecule #4: US27
| Supramolecule | Name: US27 / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #5 |
|---|---|
| Source (natural) | Organism:   Human betaherpesvirus 5 Human betaherpesvirus 5 |
| Molecular weight | Theoretical: 40 KDa |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.283836 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GCTLSAEDKA AVERSKMIDR NLREDGEKAA REVKLLLLGA GESGKSTIVK QMKIIHEAGY SEEECKQYKA VVYSNTIQSI IAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD R IAQPNYIP ...String: GCTLSAEDKA AVERSKMIDR NLREDGEKAA REVKLLLLGA GESGKSTIVK QMKIIHEAGY SEEECKQYKA VVYSNTIQSI IAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD R IAQPNYIP TQQDVLRTRV KTTGIVETHF TFKDLHFKMF DVGGQRSERK KWIHCFEGVT AIIFCVALSD YDLVLAEDEE MN RMHESMK LFDSICNNKW FTDTSIILFL NKKDLFEEKI KKSPLTICYP EYAGSNTYEE AAAYIQCQFE DLNKRKDTKE IYT HFTCAT DTKNVQFVFD AVTDVIIKNN LKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.728152 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GPGSSGSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: GPGSSGSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.432554 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: ASNNTASIAQ ARKLVEQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASENPF REKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Antibody fragment scFv16
| Macromolecule | Name: Antibody fragment scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.340482 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KGSLEVLFQ |
-Macromolecule #5: G-protein coupled receptor homolog US27
| Macromolecule | Name: G-protein coupled receptor homolog US27 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human cytomegalovirus Human cytomegalovirus |
| Molecular weight | Theoretical: 44.451742 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDMTT STNNQTLTQV SNMTNHTLNS TEIYQLFEYT RLGVWLMCIV GTFLNVLVIT TILYYRRKKK SPSDTYICNL AVADLLIVV GLPFFLEYAK HHPKLSREVV CSGLNACFYI CLFAGVCFLI NLSMDRYCVI VWGVELNRVR NNKRATCWVV I FWILAVLM ...String: DYKDDDDMTT STNNQTLTQV SNMTNHTLNS TEIYQLFEYT RLGVWLMCIV GTFLNVLVIT TILYYRRKKK SPSDTYICNL AVADLLIVV GLPFFLEYAK HHPKLSREVV CSGLNACFYI CLFAGVCFLI NLSMDRYCVI VWGVELNRVR NNKRATCWVV I FWILAVLM GMPHYLMYSH TNNECVGEFA NETSGWFPVF LNTKVNICGY LAPIALMAYT YNRMVRFIIN YVGKWHMQTL HV LLVVVVS FASFWFPFNL ALFLESIRLL AGVYNDTLQN VIIFCLYVGQ FLAYVRACLN PGIYILVGTQ MRKDMWTTLR VFA CCCVKQ EIPYQDIDIE LQKDIQRRAK HTKRTHYDRK NAPMESGEEE FLLSRGAAHH HHHHHH UniProtKB: G-protein coupled receptor homolog US27 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
Details: additives: 0.01 % w/v fluorinated octyl maltoside or 0.05 % w/v digitonin | ||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa / Details: 15 mA | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: LEICA EM GP / Details: 3 s blotting before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 3 / Number real images: 12233 / Average electron dose: 73.0 e/Å2 Details: The data from the three grids were also used to reconstruct the OCL-state US27-Gi-scFv16 complex (7RKY). |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 57624 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.0 µm / Nominal defocus min: -0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7rkx: |
 Movie
Movie Controller
Controller














































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)